ABSTRACT
Chronic myeloid leukemia(CML) is a myeloproliferative neoplasm (MPN) characterized by the presence of the Philadelphia chromosome (BCR:ABL), and there is a higher risk in untreated CML to be transformed to blast crisis phase. The development of tyrosine kinase inhibitor (TKI) imatinib was a remarkable step in CML treatment. However, patients showed resistance due to some mutations including T315I which still be the most challenging mutation in CML. Therefore, the aim of our study is to screen T315I in our Egyptian CML patients to predict and determine the frequency of T315I in patients with TKIs resistance. AS-PCR performed on 50 CML patients resistant to the first-line TKIs and 50 CML patients respond to the first-line TKIs. Positive-T315I was found in 15 (30%) of TKIs resistant cases. T315I was significantly associated with higher WBC, platelet count, cellularity of BM aspiration and biopsy when compared to those with negative-T315I. Resistant positive-T315I was significantly associated with lower cumulative OS 55.9% and shorter mean OS 93.4 months when compared to resistant negative-T315I. We conclude that T315I can exist at the diagnosis before starting treatment. Therefore, screening test for T315I can be performed at the time of diagnosis to help in choosing the suitable treatment protocol.
KEYWORDS:
Introduction
Chronic myeloid leukemia (CML) has been recognized as a clonal myeloproliferative disease and characterized by reciprocal translocation t(9; 22)(q34; q11), leading to a fusion of BCR:ABL gene called the Philadelphia (Ph) chromosome [Citation1,Citation2]. CML affects about one individual per 100,000 populations per year, with a median age of approximately 60 years at diagnosis with a slight male predominance [Citation3]. The untreated CML can transform to the blast crisis phase [Citation4].
Knowing that BCR:ABL fusion gene is a constitutively activated tyrosine kinase, tyrosine kinase inhibitors (TKIs) became a standard treatment as a targeted drugs to inhibit the tyrosine kinase activity of BCR:ABL for patients with CML [Citation5]. Imatinib was the first-generation TKI to treat CML. But despite the success on improving survival and obtaining a complete response in many patients, about 30% of patients had a resistance to the drug due to the development of mutations in the BCR:ABL oncogene [Citation6]. For that reason, second-generation TKIs (Dasatinib, Nilotinib, Bosutinib) have been developed to overcome the resistance to imatinib. All these TKIs are efficient against many mutations except the T315I mutation which considered as the most common mutation in resistance TKIs CML patients [Citation7].
The generation of T315I arises from the substitution of threonine to isoleucine at codon 315 of ABL protein leading to a modification of the structure of the binding site and eliminating the hydrogen bond between TKIs and the ABL domain [Citation8]. Therefore, ponatinib have been developed as a third generation TKI as a potent drug to overcome all kinase domain mutations but specifically to inhibit the T315I mutation. Even though ponatinib showed a high rate of response, but it also showed a high prevalence of cardiovascular events associated with the dose of 45 mg. As a result, a reduction of dose is recommended during the treatment with ponatinib. Meanwhile, various clinical trials work on many new promising drugs to inhibit the BCR:ABL mutations without causing significant side effects and toxicity [Citation9].
Therefore, the aim of our study is to screen T315I mutation in our chronic myeloid leukemia patients and its relation to TKIs resistance.
Patients and methods
The presented study is a case-control cross-sectional study, conducted on patients’ samples collected at Oncology Centre Mansoura University. It illustrated a group of 50 patients with CML resist to the first line of TKIs (Imatinib 400 mg QD), and a group included 50 CML patients who were responders to first line of TKIs (Imatinib 400 mg QD) served as a control group. All samples were obtained in accordance with the Declaration of Helsinki with informed consent from the patients, parents or guardians and approval from the Faculty of Medicine Mansoura University institutional review board (IRB code no. MS.19.02.472). In our study, the included subjects were adult patients of both sex (18–70 years) and fitting the diagnostic criteria of CML according to WHO criteria 2016 [Citation10]. The patients were excluded if associated with major cardiac or liver disease or atypical CML.
The genomic DNA was isolated from 2 ml EDTA-blood samples using QIAamp DNA blood mini kit (Qiagen). The Allele-Specific polymerase chain reaction (AS-PCR) technique has been used in our study to amplify the genomic DNA. Each sample was amplified into two tubes (one for the internal control reaction and the other for the mutant reaction). The amplified reaction of a total 25 μL included 1 μL of DNA mixed with 12.5 μL of HotStart Master Mix (Qiagen), 0.1 μL of reverse primer: 5’-GGA TGA AGT TTT TCT TCT CCA G-3’, 0.1 μL of forward control primer: 5’-GCC CCC GTT CTA TAT CAT CAC-3’ for internal control reaction OR 0.1 μL of forward specific: 5’-CCC GTT CTA TAT CAT CAT-3’ for the mutant reaction, and the reaction completed with 11.3 μL of nuclease free water (primers were designed according to Roche-Lestienne Catherine et al. [Citation11]). Amplification performed on Arkitik thermal cycler (Thermo Sientific) with the conditions: 95◦C for 15 min followed by 35 cycles of denaturation at 95◦C for 45 sec, annealing at 51.5◦C for 45 sec and extension at 72◦C for 1 min, then one cycle of a final extension at 72◦C for 8 min. The amplified products were separated by electrophoresis in 2% agarose gels with Ethidium bromide and then were visualized by UV transilluminator to detect a 158-bp band as a wild band in case of internal control reaction and as a mutant band in case of mutant reaction.
Statistical analysis
The collected data was revised, coded and tabulated using statistical package for Social Science (IBM Corp. Released 2017. IBM SPSS Statistics for Windows, Version 25.0. Armonk, NY: IBM Corp.). Data were presented and suitable analysis was done according to the type of data obtained for each parameter. p values ≤ 0.05 were considered significant.
Results
Clinico-pathological and laboratory assessments were assessed in all studied cases (). In both groups, 50% had fever, 64% had bone pain, and 12% had bleeding at the time of diagnosis. Splenomegaly did not differ significantly between responder and resistant groups, while CML resistant to TKIs had significantly higher frequency of hepatomegaly when compared to responder group (p = 0.046). CML cases resistant to TKIs had significantly higher WBC when compared to that responder to TKIs. Otherwise, laboratory data did not differ significantly between the studied CML groups. Higher blasts were significantly associated with resistant group. Sokal risk score [Citation12] did not differ significantly between TKIs responder and resistant CML groups, while higher Hasford risk score [Citation13] was significantly associated with resistant group (p < 0.001). CML resistant cases were significantly associated with higher frequency transformation when compared to responder cases (p = 0.027).
Table 1. Clinical, and laboratory data of studied CML groups.
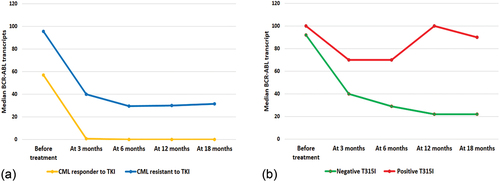
CML resistant cases had significantly higher BCR:ABL transcripts at 0, 3, 6, 12, and 18 months when compared to responder cases (). BCR:ABL transcripts decreased in the responder CML cases at 6, 12 months, and could not be detected by the 18 months in most responder cases. BCR:ABL transcripts decreased significantly across time (p < 0.001), while resistant cases had a significant decrease at 0, 3, 6 months, then remained nearly constant till 18 months.
Figure 1. Molecular monitoring of BCR-ABL transcripts. (a): BCR-ABL transcript level in the studied CML groups. (b): BCR-ABL transcript level according to T315I mutation in TKIs resistant CML.
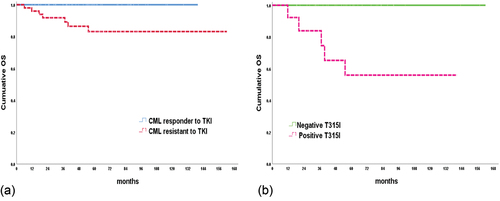
By the end of the study, all responder cases were alive, 86% of resistant cases were stationary, while 4% lost follow-up and 10% dead, with a significant difference between responder and resistant cases regarding outcome (p = 0.023). Cumulative overall survival (OS) in responder CML cases was 100%, while it was 86.7% in resistant cases with a significant difference between both groups (p = 0.008) ()
Figure 2. Cumulative overall survival. (a): Represent the OS in the studied CML groups. (b): Illustrate the OS according to T315I mutation in CML resistant to TKIs cases.
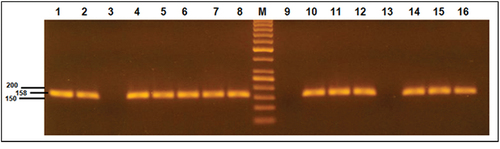
T315I positive mutation was found in 15 (30%) of TKIs resistant cases, while it was negative in all TKIs responders with p value < 0.001. illustrated the gel electrophoresis results of T315I mutation.
Figure 3. Agarose gel electrophoresis results of T315I mutation. Lane M is 50 bp DNA marker. Lanes 1, 5,7,11 and 15 are positive T315I mutation. Lanes 3, 9 and 13 are negative T315I mutation cases. Lanes 2,4,6,8,10,12,14 and 16 represent the wild type of all cases.
We divided the resistant group into two subgroups, one with negative T315I mutation and the other one with positive T315I mutation (). Fever was significantly associated with positive T315I mutations. Other clinical parameters were not significantly associated with T315I mutation. Also, T315I positive group was significantly associated with higher WBC, platelet count, cellularity of BM when compared to those with negative T315I. No significant association was found between positive and negative T315I resistant CML cases regarding disease phases.
Table 2. Clinical and laboratory data of CML resistant cases according to T315I mutation.
BCR:ABL transcript was higher in positive T315I cases at all time points. This increased level did not reach significant level by 0, 3rd, 6th months, but reached by the 12th (p 0.007) and 18th (p 0.012) months. It is noticed that BCR:ABL transcript in negative T315I cases decreased significantly by time (p 0.001), although it was detected till 18th month (). By the end of the study, all negative T315I resistant cases still alive. On the other hand, 53.3% of the positive cases with T315I alive and 33.3% died. Resistant positive T315I group was significantly associated with lower cumulative OS (55.9% versus 100%) and shorter mean OS (93.4 versus 169 months) when compared to resistant negative T315I group ().
Logistic regression analysis was conducted for prediction of TKI resistance in CML cases using LDH, blasts, BCR:ABL transcript, disease phase, Hasford score, T315I as confounders (). Higher BCR:ABL transcript before treatment, Hasford score and positive T315I were associated with TKI resistance in univariable analysis. However, in multivariable analysis, only Hasford score and positive T315I were suggested to be independent predictors of TKI resistance in CML cases.
Table 3. Logistic regression analysis for prediction of TKI resistance.
Lastly, Cox regression analysis was conducted for prediction of OS in CML cases, using marrow blasts, LDH, BCR:ABL transcript before treatment, disease phase, Hasford score and T315Imutation as covariates. Higher BM blasts, Hasford score and positive T315I mutations were suggested to be independent risk predictors for shorter OS in CML cases in uni and multivariable analyses ().
Table 4. Cox regression analysis for prediction of OS in CML cases.
Discussion
The discovery of TKIs was a remarkable step in the treatment of CML and in monitoring the BCR:ABL level, but the challenge was risen when several mutations were found to be responsible for the resistance to TKIs especially the T315I mutation. Therefore, the screening of T315I mutation had been recommended to help in the therapeutic decision for CML patients. Therefore, we represent a detection method of T315I mutation using allele specific PCR (AS-PCR) technique for screening our patients in the Oncology Center Mansoura University, Egypt.
In our study, T315I positive mutations were found in 15 (30%) of TKI resistant patients, while the remaining 35 (70%) of the resistant cases were negative T315I mutation. This high frequency of positive T315I is in the same as the frequency reported from patients in Pakistan and China with 37.5%, 22.9% respectively [Citation14,Citation15], but it is not as those reported from patients in other populations such as Iraq and Iran that showed a lower existing of T315I with 3% and 7% respectively [Citation6,Citation16]. These differences may relate to the time whether the T315I screening was done at the beginning of diagnosis or after noticing the resistance to the TKI, also related to the stage of the disease at which the mutation was detected whether in chronic phase or in advanced phase, also the patient’s ethnic origin for each study. In our study, only two cases with positive T315I were at accelerated phase while the other 13 positive T315I were at chronic phase. This finding suggesting that the existence of T315I mutation has no relation to the stage of disease and this is in controversy to different study mentioned that T315I mutation can be detected more commonly in advanced phases of disease [Citation17].
T315I mutation in our study has been correlated with some clinical and laboratory data (fever, WBC, and platelet count) with p value 0.03, 0.003, <0.001 respectively. On the other hand, El-Menoufy et.al, and Rejali & Poopak reported no significant relationship between the presence of T315I mutation and patient’s clinical signs [Citation18,Citation19]. CML resistant cases were significantly associated with higher frequency transformation when compared to responder cases (p = 0.027). Moreover, by the end of our study, all negative T315I resistant cases still alive and only about half of positive T315I resistant cases still alive (53.3%) while about one third of mutated cases died (33.3%).
The increased levels of BCR:ABL transcript in resistant cases have been found to be correlated with the presence of T315I (median 90) when compared with negative T315I (median 22). Positive T315I was significantly associated with lower cumulative OS (55.9% versus 100%) and shorter mean OS (93.4 versus 169 months) when compared to CML with negative T315I which indicate the additive effect of poor prognosis by the presence of T315I mutation. This agrees with a study reported a reduced OS and a significant poor prognosis associated with the presence of T315I [Citation20,Citation21], and in controversy with El-Menoufy et al. and Jabbour et al. who demonstrated no significant differences in survival [Citation18,Citation22].
From these results, we conclude that T315I mutation can exist at the diagnosis before starting treatment. Therefore, screening test as AS-PCR for T315I can be performed at the time of diagnosis to help in choosing the suitable treatment protocol for CML patients.
Acknowledgments
The authors like to acknowledge all participants in this work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Zhang J, Jin Y, Pan J. Inhibitory effect of the anthelmintic drug pyrvinium pamoate on T315I BCRABLpositive CML cells. Mol Med Rep. 2017;16(6):9217–9223. doi: 10.3892/mmr.2017.7685
- Johansson B, Fioretos T, Mitelman F. Cytogenetic and molecular genetic evolution of chronic myeloid leukemia. Acta Haematol. 2002;107(2):76–94. doi: 10.1159/000046636
- Ning L, Hu C, Lu P, et al. Trends in disease burden of chronic myeloid leukemia at the global, regional, and national levels: a population-based epidemiologic study. Exp Hematol Oncol. 2020;9(1):1–4. doi: 10.1186/s40164-020-00185-z
- Khorshied MM, Shaheen IA, Khalil RE, et al. Methylene tetrahydrofolate reductase (MTHFR) gene polymorphisms in chronic myeloid leukemia: an Egyptian study. Med Oncol. 2014;31(1):794. doi: 10.1007/s12032-013-0794-2
- Braun TP, Eide CA, Druker BJ. Response and resistance to BCR-ABL1-targeted therapies. Cancer Cell. 2020;37(4):530–42. doi: 10.1016/j.ccell.2020.03.006
- Dhahi MA, Matti BF, Fadel S. Molecular screening for T315I and F317L resistance mutations in Iraqi chronic myeloid leukemia non-responders patients to imatinib. Cancer Clin Oncol. 2013;2(2):55–61. doi: 10.5539/cco.v2n2p55
- Lussana F, Intermesoli T, Stefanoni P, et al. Mechanisms of resistance to targeted therapies in chronic myeloid leukemia. Mechanisms of drug resistance in cancer therapy. Handb Exp Pharmacol. 2018;249:231–250.
- Soverini S, Colarossi S, Gnani A, et al. Contribution of ABL kinase domain mutations to imatinib resistance in different subsets of philadelphia-positive patients: by the GIMEMA working party on chronic myeloid leukemia. Clin Cancer Res. 2006;12(24):7374–9. doi: 10.1158/1078-0432.CCR-06-1516
- Sampaio MM, Santos ML, Marques HS, et al. Chronic myeloid leukemia-from the Philadelphia chromosome to specific target drugs: a literature review. World J Clin Oncol. 2021;12(2):69. doi: 10.5306/wjco.v12.i2.69
- Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–405. doi: 10.1182/blood-2016-03-643544
- Roche-Lestienne C, Soenen-Cornu V, Grardel-Duflos N, et al. Several types of mutations of the abl gene can be found in chronic myeloid leukemia patients resistant to STI571, and they can pre-exist to the onset of treatment. Blood. 2002;100(3):1014–8. doi: 10.1182/blood.V100.3.1014
- Sokal JE, Cox EB, Baccarani M, et al. Prognostic discrimination in“good-risk” chronic granulocytic leukemia. Blood. 1984;63(4):789–799. doi: 10.1182/blood.V63.4.789.789
- Hasford J, Pfirrmann M, Hehlmann R, et al. A new prognostic score for survival of patients with chronic myeloid leukemia treated with interferon alfa writing committee for the collaborative CML prognostic factors project group. J Natl Cancer Inst. 1998;90(11):850–9. doi: 10.1093/jnci/90.11.850
- Iqbal Z, Aleem A, Iqbal M, et al. Sensitive detection of pre-existing BCR-ABL kinase domain mutations in CD34+ cells of newly diagnosed chronic-phase chronic myeloid leukemia patients is associated with imatinib resistance: implications in the post-imatinib era. PloS One. 2013;8(2):e55717. doi: 10.1371/journal.pone.0055717
- Liu J, Yang H, Xu X, et al. Mutations in the BCRABL1 kinase domain in patients with chronic myeloid leukaemia treated with TKIs or at diagnosis. Oncol Lett. 2020;20(2):1071–1076. doi: 10.3892/ol.2020.11650
- Chahardouli B, Zaker F, Mousavi SA, et al. Evaluation of T315I mutation frequency in chronic myeloid leukemia patients after imatinib resistance. Hematology. 2013;18(3):158–62. doi: 10.1179/1607845412Y.0000000050
- Willis SG, Lange T, Demehri S, et al. High-sensitivity detection of BCR-ABL kinase domain mutations in imatinib-naive patients: correlation with clonal cytogenetic evolution but not response to therapy. Blood. 2005;106(6):2128–37. doi: 10.1182/blood-2005-03-1036
- El-Menoufy MA, El Naggar AA, Ziada LE. ‘Gatekeeper’ mutation in patients with chronic myeloid leukemia resistant to imatinib therapy: effect on survival. Egypt J Haematol. 2018;43(3):138. doi: 10.4103/ejh.ejh_18_18
- Rejali L, Poopak B, Hasanzad M, et al. Characterizing of four common BCR-ABL kinase domain mutations (T315I, Y253H, M351T and E255K) in Iranian chronic myelogenous leukemia patients with imatinib resistance. Iran J Cancer Prev. 2015;8(3). doi: 10.17795/ijcp2334
- Kuntegowdanahalli LC, Kanakasetty GB, Thanky AH, et al. Prognostic and predictive implications of Sokal, Euro and EUTOS scores in chronic myeloid leukaemia in the imatinib era—experience from a tertiary oncology centre in Southern India. Ecancermedicalscience. 2016;10. doi: 10.3332/ecancer.2016.679
- Nicolini FE, Ibrahim AR, Soverini S, et al. The BCR-ABLT315I mutation compromises survival in chronic phase chronic myelogenous leukemia patients resistant to tyrosine kinase inhibitors, in a matched pair analysis. Haematologica. 2013;98(10):1510. doi: 10.3324/haematol.2012.080234
- Jabbour E, Kantarjian H. Chronic myeloid leukemia: 2016 update on diagnosis, therapy, and monitoring. Am J Of Hematol. 2016;91(2):252–265. doi: 10.1002/ajh.24275
