ABSTRACT
Mutations in the KRAS oncogene at codons 12, 13, and 22 of exon 2 are common in colorectal cancer (CRC). These mutations lead to constitutive RAS pathway activation and are thought to promote tumor progression. However, their correlations with clinical features remain uncertain. KRAS exon 2 mutations were analyzed in 173 (CRC) patients by polymerase chain reaction (PCR) and sequencing. Associations with demographic and pathological factors were examined. Three specific mutations were identified: G12C, G13D, and G22E. 144 patients had KRAS mutations while 29 were wild type. Mutations were more frequent vs wild type (83% vs 17%, p = 0.0001). Mutated tumors were associated with ages ≤ 40 vs ages ˃ 40 (86% vs 82%, p = 0.038), female gender (90% vs 80%, p = 0.007), and moderate differentiation (95% vs 77%, p = 0.033). KRAS exon 2 mutations were prevalent in this cohort and associated with clinicopathologic features. The results suggest these mutations increase colorectal cancer risk and support further investigation of their prognostic utility.
Introduction
Colorectal cancer (CRC) is a leading contributor to cancer-related deaths across the globe, ranking as the third most prevalent form of cancer internationally. The growth of (CRC) is propelled by the accumulation of genetic and epigenetic changes that facilitate the initiation and advancement of tumors [Citation1]. Many studies have shown that colorectal cancer is linked to S. Gallolyticus infection, but other studies have indicated that this association still needs further clarification [Citation2]. The Kirsten rat sarcoma (KRAS) is a commonly mutated oncogene in (CRC), affecting 30–50% of cases. It encodes a guanosine triphosphatase (GTPase) that plays a crucial role in regulating cellular signaling pathways that control cell growth, differentiation, and survival. Mutations in the KRAS gene can increase the risk of developing malignant tumors in various organs such as the intestine, rectum, lung, pancreas, colorectal, and urogenital system [Citation3,Citation4]. Oncogenic mutations in KRAS lead to constitutive activation of downstream pathways, enhancing cancer cell growth and survival [Citation5]. The majority of KRAS mutations in (CRC) occur in codons 12 and 13 of exon 2, resulting in single amino acid substitutions that impair GTPase activity [Citation6]. Specific KRAS mutations have been associated with resistance to anti-epidermal growth factor receptor (anti-EGFR) therapies in metastatic (CRC) [Citation7]. However, the relationship between KRAS mutation status and clinical or histopathologic features remain incompletely understood. Some studies have suggested associations of KRAS mutations with factors like age, sex, tumor location, differentiation, and microsatellite instability, while others have found no significant correlations [Citation8]. The prevalence of the KRAS G12D mutation was observed in both pancreatic and CRC, with no correlation to sex, age, and tumor grade, while in lung cancer, KRAS gene mutations were significant in males. In addition, these mutations were associated with right colon cancer but not the left, as is the case with BRAF, PIK3CA and p53 gene mutations [Citation9–11]. Other studies have shown no difference based on the location and spread of the tumor, KRAS G12C, G13D, and G22E mutations were more prevalent and likely to be associated with microsatellite instability in CRC [Citation12]. Detailed characterization of KRAS mutations in exon 2 and analysis of potential correlations with clinicopathologic variables may provide insights into the utility of KRAS status for prognosis or prediction of treatment response. This study performed a mutational analysis of KRAS exon 2 in (CRC) patients and investigated associations with demographic and tumor histology factors like age, sex, tumor location, differentiation
Material and methods
Study group
A group of 173 patients with (CRC) was selected from Medical City Hospital in Baghdad to study the alterations in the KRAS gene. The clinical details, including age, sex, and tumor characteristics, were collected from the patients’ medical records after obtaining consent from the patients and hospital administration. In this study, the selection criteria for patients were based on ensuring that they had CRC and had a complete information record in the hospital, while patients who were unable to visit the hospital periodically for treatment and who had a second disease were excluded. All patients provided written informed consent, and this study was approved by the Ethical Committee of Baghdad Teaching Hospital, College of Medicine, Baghdad, Iraq (BMI 63).
Genomic DNA extraction
The DNA extraction process from blocks tissue samples that were fixed in formalin and embedded in paraffin (FFPE) was carried out using Qiagen kit (QIAamp DNA FFPE tissue kit, a German company) according to the manufacturer’s instructions. The isolated DNA was stored at −80°C for the purpose of completing the KRAS sequence analysis program. The DNA quality and concentration ratio were 1.7–5 at the wavelength of 260/280 nm, confirming the purity of the DNA. The concentration was measured using a NanoDrop™ spectrophotometer (Nanodrop Technologies, Inc).
The polymerase chain reaction (PCR)
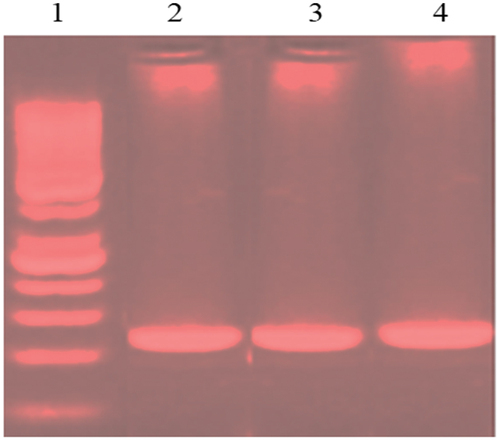
The Polymerase Chain Reaction (PCR) amplification was used to generate a product (263bp) specific to exon 2 of KRAS gene; utilizing specific primers (forward, 5´ GGTGAGTTTGTATTAAAAGGTACTGG; and reverse, 5´-TCCTGCACCAGTAATATGCA). The kit and primers have been explained and described previously [Citation13]. Than the reactions were performed in a volume of 20 μl, consisting of 5 μl of DNA template, 0.5 μl of forward inner primer, 0.5 μl of reverse inner primer,10 μl green master mix (Promega/USA) and 4 μl nuclease free water. Than the cycles were 49, initial denaturation at 95°C for 3 min, followed by denaturation at 95°C for 15 s, annealing at 55°C for 15 s, and extension at 72°C for 15 s. The agarose 1.5% with 0.5 μl of ethidium bromide was used to examine the PCR products ().
KRAS DNA sequencing
All samples with primers were sent to Macrogen Company (USA) to complete the sequencing reaction. Than the PCR products of the KRAS gene were sequenced in both reverse and forward primers using directions the Big Dye Terminator v3.1 cycle sequencing kit on ABI 3730 (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. The results were analyzed using Mutation Surveyor software (Mega-11), and compared with the information in the Gen Bank database of the National Center for Biotechnology Information (NCBI) for the standard KRAS gene in order to identify mutations. This analysis allows for the identification of genetic mutations throughout the genome identified by PCR ().
Statistical analysis
To determine the effect of variables such as age, gender, and histological condition of the (CRC) cancer patients and the wild type, as well as the influence of these variables on the mutation status of codons 12, 13, and 22. The data were summarized as percentages and frequencies. The Statistical Analysis System (SAS) program (2018) was used to study statistical significance. Percentages were compared using the chi-square test with a significance level of 0.05 and a probability of 0.01 [Citation14].
Results
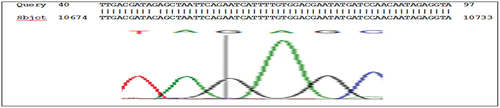
The results of the sequencing analysis for mutations in the KRAS gene, exon 2 showed the presence of mutations in codon 12: GGT>TGT substitution G>T, (rs121913530), codon 13: ggc>gac substitution G>A, (rs397517040), and codon 22: cag>gag substitution C>G ( , and respectively).
Figure 3. g.10653 codon 12 substitution G>T, ggt> tgt Rs121913530 nucleotide sequence (forward) in KRAS gene, exon 2.
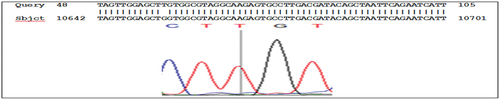
Figure 4. g.10658 codon 13 substitution G>A, rs397517040 ggc>gac nucleotide sequence (forward) in KRAS, exon 2.
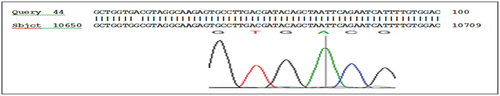
Figure 5. g.10683 codon 22 cag>gag substitution C>G, nucleotide sequence (forward) in KRAS gene, exon 2.
Distribution the KRAS gene with (CRC)
There were 173 samples of (CRC) documented, of which 144 (83.2%) had KRAS mutations and 29 (16.8%) were KRAS wild-type. The age, sex, and cell histological findings were compared between the KRAS gene mutants and the wild type, and these results were documented. KRAS mutations were more frequent in younger patients, 86.5% of patients ≤40 years old had mutations, compared to 82% of patients >40 years old. This difference was statistically significant (p = 0.0388). There was a significant difference in KRAS mutation frequency by gender, 90.5% in females vs. 80% in males (p = 0.0074). KRAS mutations were more frequent in moderately differentiated histological cell types (95%), compared to well-differentiated (66.7%) and poorly differentiated (76%). This difference was statistically significant (p = 0.0327). In summary, this data shows KRAS mutations are very common in (CRC), and the mutation frequency differs by age, gender and histological cell type. This study found higher mutation rates in younger patients, females, and moderately differentiated tumors. This provides useful epidemiological insights into the distribution of KRAS mutations in (CRC) ().
Table 1. Distribution the KRAS gene with (CRC) study in wild-type and mutation according to age, gender and histological-cell.
Correlation between age, gender and histological cell with KRAS mutation
A statistically significant association was found between age and KRAS mutation status (p = 0.0054). Patients ≤40 years old had a higher rate of codon 13 mutations (51.1%), while patients ˃ 40 years old had a slightly lower rate (50.5%). Gender was found to have a significant association with KRAS gene mutation status (p = 0.0006). Males had a higher rate of codon 13 mutations (60.4%), while females had a rate of codon 12 mutations (48%). The presence of a KRAS mutation is significantly correlated with the histological cell type (p = 0.0015). Differentiated tumors had a higher rate of codon 12 mutation (68.8%) and moderately-differentiated of codon 13 mutation (64.4%). Overall, codon 13 mutations appear to be more common and frequent in younger patients, males, and moderate-cell tumors based on this data. Codon 13 and 12 mutations are most frequent overall. The Chi-square test shows there are statistically significant associations between these clinicopathological features and KRAS mutation status. This suggests KRAS mutations may correlate with specific phenotypic or clinical characteristics ().
Table 2. Correlation between age, gender and histological cell with KRAS mutation.
Codon 12, codon 13 and codon 22 and amino acid change
In , the 38.2% (55/144) of cases had a mutation in KRAS codon 12, specifically a GGT>TGT mutation leading to a Glycine to Cysteine amino acid change. 50.7% (73/144) of cases had a mutation in KRAS codon 13, specifically a GGC>GAC mutation leading to a Glycine to Aspartic Acid change. 11.1% (16/144) of cases had a mutation in KRAS codon 22, specifically a CAG>GAG mutation leading to a Glycine to Glutamic Acid change. The codons 12 and 13 mutations were significantly more common than the codon 22 mutation based on the chi-square test results showing p < 0.0001. Overall, this data shows codons 12 and 13 are hot spots for KRAS mutations in this (CRC) cohort, with the GGT>TGT and GGC>GAC changes being the most prevalent. The codon 22 CAG>GAG mutation was less common. Testing for these common KRAS codons 12, 13 and 22 mutations would be important for guiding anti-EGFR therapy in (CRC) patients from this population.
Table 3. Codons 12, 13 and 22 and amino acids change.
Discussion
Investigating the correlation between mutations in the KRAS gene and factors such as age, gender, and cell histology
In this study, mutations in the KRAS gene occurred more frequently than the wild-type version, 83.2%, 16.8% respectively (p-value = 0.0001), this may be related the race of a population. In cecal site cancer, the mutant KRAS gene recorded a significant difference compared to the wild type (24% vs. 12%, respectively; p < 0.0001) [Citation15]. While other studies have shown that the prevalence of KRAS gene mutations in patients with (CRC) was 38%-50% [Citation16,Citation17]. No significance was documented regarding tumor status between the mutant KRAS gene was 40 cases and the wild type was 26 cases [Citation18]. This study showed that age ≤40 years old develops (CRC) more frequently than those over 40 years old. This aligns with previous studies that also reported higher rates of KRAS mutations in early-onset (CRC) [Citation19]. No link between KRAS mutation prevalence and age [Citation9]. The prevalence of KRAS mutation in (CRC) is higher in older patients compared to younger patients [Citation10]. This study also indicated that females are significantly more susceptible to (CRC) than males. There is a higher frequency of KRAS mutation in females compared to males in (CRC) [Citation16]. As age increased, there was a higher number of colorectal polyps observed in females compared to males [Citation20]. The male higher frequency than female in (CRC) [Citation17]. The study conducted a meta-analysis and found no significant relationship between sex and KRAS gene mutations in (CRC) [Citation21]. Histologically, the KRAS gene mutations were more common with moderately differentiated cell types compared to well-differentiated and poorly differentiated cell types. Another study found that KRAS mutations were more prevalent in the moderate-cell type compared to the well-cell and differentiation-cell types in (CRC) [Citation22]. There was no significant difference in the prevalence of KRAS mutations between the well-differentiated and moderately differentiated cell types in (CRC) [Citation23]. The prevalence of KRAS mutations was higher in the poorly differentiated cell type compared to the well-differentiated and moderately differentiated cell types in (CRC) [Citation9]. Most studies have confirmed the existence of significant correlations between KRAS mutations and factors such as tumor grade, differentiation, age, gender, and microsatellite instability [Citation10,Citation16,Citation20], while some sources have linked the harmful consequences of KRAS gene mutations with response to treatment and survival [Citation24,Citation25]. The results of this study show the interaction between KRAS gene mutations and their effect on clinicopathological characteristics. Which may provide predictive insights into the distribution of KRAS mutations in CRC, disease progression and survival.
Analysis mutations of codons 12, 13 and 22
This research has verified that mutations occur more often in codon 13 than in codons 12 and 22 among individuals age ≤ 40. This is consistent with some previous studies; the higher rates of codon 12 mutations in patients < 50 years old [Citation24]. The prevalence of KRAS mutation in codon 12 is higher (84%) in (CRC) patients <30 years old compared to age ≥30 years old (19%) with codon 13 mutation [Citation25]. In males, the presence of KRAS codon 13 mutation is considered morally significant in 60.4% of cases, whereas in females, the presence of codon 12 mutation is morally significant in 48% of cases. This is consistent with previous studies [Citation15]. There is a higher prevalence of codon 12 mutations in females [Citation26]. Both male and female individuals showed a high prevalence (83%) of KRAS mutation in codon 12, while codon 13 was only observed in a smaller proportion (17%) of both genders [Citation27]. In Histological-cell, there was a significant mutation of the KRAS gene at codon 12 and 13 in the Differentiation-cell and Moderate-cell, respectively. The prevalence of the G12C mutation was more common in a well-cell and differentiation-cell at a rate of 44.7% and 68.8% respectively, while the G13D mutation was more common in a moderate-cell at a rate of 64.4%. The G12C mutation may be a cause of tumor emergence, while the G13D mutation then the G12C mutation reasons for increased tumor aggressiveness. Results of this study is consistent with other studies that have documented that these mutations play distinct roles in the different stages of tumor development, poor prognosis, decreased survival rate, and decreased benefit from EGFR therapy [Citation12,Citation20,Citation28]. In the Differentiation-cell the G22E (18.7%), while the G13D (12.5%) this confirms the association of the G22E mutation with tumor behavior and increased aggressiveness and spread of CRC, and this is consistent with other studies that documented this mutation in advanced tumors [Citation29]. Other studies have shown that KRAS G12C, G13D, and G22E mutations may be associated with the induction of persistent dysregulated signaling through the RAS/MAPK pathway, which may contribute to the development and progression of CRC [Citation4,Citation5,Citation29]. This is consistent with the results of this study: the importance of KRAS mutations, especially G12C G13D G22E, in tumor proliferation, development, and progression. This emphasizes the need for targeted therapies to address the risks of these mutations associated with tumor growth and development. A study found that codons 12 and 13 had a high prevalence of well-modified forms, at 95%. However, the prevalence of poorly modified forms in codons 12 and 13 was low, at 5% and 8% respectively [Citation11]. There were no significant levels of mutations found in codons 12 and 13 of the KRAS gene in the histologic cell case [Citation30].
Investigation of the amino acid alterations within codons 12, 13, and 22
The most frequent mutations are found in codons 12 and 13, while codon 22 mutations are less common [Citation31,Citation32]. This is consistent with this study. This data shows codon 13 mutations (GGC>GAC; Gly > Asp) occurring at the highest frequency of 50.7%. This aligns with previous research indicating codon 13 mutations represent about 50% of all KRAS mutations in (CRC) [Citation31]. The Gly > Cys mutation in codon 12 (GGT>TGT) occurred in 38.2% of cases. Moreover, this is also consistent with the literature, as codon 12 mutations account for approximately 35–45% of KRAS mutations [Citation33]. In a separate research conducted on patients with (CRC), it was observed that out of the samples tested, 40% had a G12D mutation and 38.6% had a G13D mutation in the KRAS gene [Citation34]. The Gly to Glu mutation in codon 22 (CAG>GAG) was least frequent at 11.1%. Previous studies have found codon 22 mutations in 0–17% of colorectal tumors [Citation28]. The significant p-value for the codon 12 mutation suggests it differed between comparison groups in this study. KRAS mutations can vary by factors like gender, tumor location, and pathological stage [Citation20]. This study is consistent with other studies that have shown that to guide treatment decisions and individualize treatment, it is recommended to test for KRAS mutations, especially at codons 12, 13, and 22, in patients with metastatic (CRC) before initiating anti-EGFR therapy [Citation12,Citation28,Citation29].
Conclusion
KRAS mutations at codons 12, 13, and 22 are consistent with previous reports written in (CRC). Codon 13 mutations were a significant difference in ages and gender groups, while codon 12 mutations showed a significant difference in Histological-cell groups. Codon 22 mutation was more prevalent than codon 13 mutations in the histological-cell groups. Generally, the oncogenic KRAS mutations were prevalent and associated with earlier disease onset, being female, and having moderately differentiated histological grades. These associations suggest that KRAS mutations promote colorectal tumor development. Analyzing mutation patterns and clinical correlations can provide insight into how mutated KRAS alleles contribute to cancer risk and progression.
Author contribution
S.B. designed and performed the experiments, analyzed them, wrote, revised, and edited the manuscript.
Ethical approval
All patients provided written informed consent, and this study was approved by the Ethical Committee of Baghdad Teaching Hospital, College of Medicine, Baghdad, Iraq (BMI 63).
Acknowledgments
Thanks and appreciation to the Institute of Genetic Engineering and Biotechnology for Postgraduate Studies/University of Baghdad for their approval to conduct this research.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Xi Y, Xu P. Global colorectal cancer burden in 2020 and projections to 2040. 2021 Oct;14(10):101174. doi: 10.1016/j.tranon.2021.101174
- Eldegla E, Abdel-Wahhab M, Moemen D. Association between streptococcus gallolyticus and colorectal cancer in Mansoura University hospitals. Egypt J Basic Appl Sci. 2021;8(1):397–406. doi: 10.1080/2314808X.2021.2001618
- Hong D, Fakih M, Strickler J, et al. KRAS G12C inhibition with sotorasib in advanced solid tumors. N Engl J Med. 2020;383(13):1207–1217. doi: 10.1056/NEJMoa1917239
- Sui X, Chen Y, Liu B, et al. The relationship between KRAS gene mutation and intestinal flora in tumor tissues of colorectal cancer patients. Ann Transl Med. 2020;8(17):1085. doi: 10.21037/atm-20-5622
- Ferreira A, Pereira F, Reis C, et al. Crucial role of oncogenic KRAS mutations in apoptosis and autophagy regulation: therapeutic implications. Cells. 2022;11(14):2183. doi: 10.3390/cells11142183
- Vitiello P, Cardone C, Martini G. Receptor tyrosine kinase-dependent PI3K activation is an escape mechanism to vertical suppression of the EGFR/RAS/MAPK pathway in KRAS-mutated human colorectal cancer cell lines. J Exp Clin Cancer Res. 2019;38(1):41. doi: 10.1186/s13046-019-1035-0
- Korkmaz L, Coskun H, Dane F. Kras-mutation influences outcomes for palliative primary tumor resection in advanced colorectal cancer-a Turkish oncology group study. Surg Oncol. 2018;27(3):485–489. doi: 10.1016/j.suronc.2018.05.032
- Gbolahan O, O’Neil B. Update on systemic therapy for colorectal cancer: biologics take sides. Transl Gastroenterol Hepatol. 2019;4:9. doi: 10.21037/tgh.2019.01.12
- Hasbullah H, Sulong S, Che Jalil A, et al. KRAS mutational profiles among colorectal cancer patients in the east coast of Peninsular Malaysia. Diagnost (Basel). 2023;13(5):822. doi: 10.3390/diagnostics13050822
- Rawla P, Sunkara T, Barsouk A, et al. Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Prz Gastroenterol. 2019;14(2):89–103. doi: 10.5114/pg.2018.81072
- Sirirat S, Paninee T, Nuntaree C, et al. Validation of a multiplex allele-specific polymerase chain reaction assay for detection of KRAS gene mutations in formalin-fixed, paraffin-embedded tissues from colorectal cancer patients. PLOS ONE. 2016;1(1):e0147672. doi: 10.1371/journal.pone.0147672
- Ana F, Vicente A, Elena A. The frequency of specific KRAS mutations, and their impact on treatment choice and survival, in patients with metastatic colorectal cancer. Oncology. 2023;28(10):e902–e909. doi: 10.1093/oncolo/oyad117
- Nikiforova M, Lynch R, Biddinger P. RAS point mutations and PAX8-PPARγ rearrangement in thyroid tumors: evidence for distinct molecular pathways in thyroid follicular carcinoma. J Clin Endocrinol Metab. 2003;88(5):2318–2326. doi: 10.1210/jc.2002-021907
- Statistical Analysis System, User’s Guide. Statistical. Version 9.6th ed. USA Cary NC: SAS Institute Inc; 2018.
- Phipps A, Buchanan D, Makar K, et al. KRAS-mutation status in relation to colorectal cancer survival: the joint impact of correlated tumour markers. Br J Cancer. 2013;108(8):1757–1764. doi: 10.1038/bjc.2013.118
- Judd J, Abdel Karim N, Khan H, et al. Characterization of KRAS mutation subtypes in non–small cell lung cancer. Mol Cancer Ther. 2021;20(12):2577–2584. doi: 10.1158/1535-7163.MCT-21-0201
- Strickler H, Yoshino T, Stevinson K. Prevalence of KRAS G12C mutation and co-mutations and associated clinical outcomes in patients with colorectal cancer: a systematic literature review. The Oncolog. 2023;28(11):e981–e994. doi: 10.1093/oncolo/oyad138
- Yi-Jian T, Sheng H, Hung L, et al. Differences in gene mutations according to gender among patients with colorectal cancer. World J Surg Oncol. 2023;21:378. doi: 10.1186/s12957-018-1431-5
- Mauri G, Sartore A, Russo A, et al. Early-onset colorectal cancer in young individuals. Mol Oncol. 2019;13(2):109–131. doi: 10.1002/1878-0261.12417
- Ameer A, Rana N, Haider A, et al. Investigate the role of PIK3CA gene expression in colorectal polyp development. Egypt J Basic Appl Sci. 2023;10(1):594–604. doi: 10.1080/2314808X.2023.2245992
- Choi Y, Kim N. Sex difference of colon adenoma pathway and colorectal carcinogenesis. World J Mens Health. 2024;42(2):256–282. doi: 10.5534/wjmh.230085
- Hayama T, Hashiguchi Y, Okamoto K. G12V and G12C mutations in the gene KRAS are associated with a poorer prognosis in primary colorectal cancer. Int J Colorectal Dis. 2019;34(8):1491–1496. doi: 10.1007/s00384-019-03344-9
- Chowdhury S, Ferdous Ara J, Mili M, et al. Mutational profile of KRAS, NRAS, BRAF, PIK3CA, and AKT1 genes in colorectal cancer patients in a tertiary care hospital, Dhaka. Adv C Biol–Metastasis. 2022;5:100054. doi: 10.1016/j.adcanc.2022.100054
- Andreyev H, Norman R, Cunningham D, et al. Kirsten ras mutations in patients with colorectal cancer: the ‘RASCAL II’ study. Br J Cancer. 2001;85(5):692–696. doi: 10.1054/bjoc.2001.1964
- Yu I, Paul L, Mai Y, et al. Analyses of clinicopathological, molecular, and prognostic associations of KRAS codon 61 and codon 146 mutations in colorectal cancer: cohort study and literature review. Mol Cancer. 2014;13(1):135. doi: 10.1186/1476-4598-13-135
- Qunaj L, May S, Neugut I, et al. Herzberg prognostic and therapeutic impact of the KRAS G12C mutation in colorectal cancer. Front Oncol. 2023;13:1252516. doi: 10.3389/fonc.2023.1252516
- Carlos G, Veronica A, Ilana Z, et al. KRAS mutations: variable incidences in a Brazilian cohort of 8,234 metastatic colorectal cancer patients. BMC Gastroenterol. 2014;14(1):73. doi: 10.1186/1471-230X-14-73
- Soukaina B, Abdelilah L, Fatima El B, et al. Clinical significance of somatic mutations in RAS/RAF/MAPK signaling pathway in Moroccan and North African colorectal cancer patients. Asian Pac J Cancer Prev. 2022;23(11):3725–3733. doi: 10.31557/APJCP.2022.23.11.3725
- Kazunori T, Motohiko T, Hiroyuki S. A novel activating mutation of the K-ras gene in human primary colon adenocarcinoma. Biochem Biophys Res Commun. 2000;278(3):653–658. doi: 10.1006/bbrc.2000.3839
- Harry H, David T, Qian S, et al. KRAS codon 12 and 13 mutations in relation to disease-free survival in BRAF –wild-type stage III colon cancers from an adjuvant chemotherapy trial (N0147 alliance). Clin Cancer Res. 2014;20(11):3033–3043. doi: 10.1158/1078-0432.CCR-13-3140
- Arrington K, Eileen H, Lee W. Prognostic and predictive roles of KRAS mutation in colorectal cancer. IJMS. 2012;13(10):12153–12168. doi: 10.3390/ijms131012153
- Mun T, Jung Y, Wan M, et al. Current advances and trends in KRAS targeted therapies for colorectal cancer. Mol Cancer Res. 2022;20(1):30–44. doi: 10.1158/1541-7786.MCR-21-0248
- Shafia R, Shimon G, Michael G, et al. Therapeutic targets of KRAS in colorectal cancer. Cancers (Basel). 2021;13(24):6233. doi: 10.3390/cancers13246233
- Choong E, Seung Y, Bonhan K, et al. Rapid and accurate detection of KRAS mutations in colorectal cancers using the isothermal-based optical sensor for companion diagnostics. Oncotarget. 2017;8(48):83860–83871. doi: 10.18632/oncotarget.20038