ABSTRACT
When scientists alter the genome of an organism, we typically reduce its ability to reproduce in the wild. This limitation has prevented researchers from rendering wild insects unable to spread disease, programing pests to ignore our crops, using genetics to precisely remove environmentally damaging invasive species, and much more. Gene drive occurs when a vertically transmitted genetic element reliably spreads through a population over generations despite providing no reproductive advantage to each host organism. Until recently, scientific efforts to take advantage of this natural phenomenon achieved only limited success. The advent of CRISPR genome editing has dramatically accelerated efforts to harness gene drive. Small groups of scientists may now be capable of unilaterally altering entire wild populations, and through them, the shared environment. Determining whether, when, and how to develop gene drive interventions responsibly will be a defining challenge of our time. Here we describe capabilities, safeguards, applications, and opportunities relevant to gene drive technologies.
But Natural Selection … is a power incessantly ready for action, and is as immeasurably superior to Man’s feeble efforts, as the works of Nature are to those of Art. – Charles Darwin
Introduction
Humanity’s efforts to harness gene drive predate our knowledge of the structure of DNA (Gould Citation2008; Burt and Trivers Citation2008). In the early 1940s, F. L. Vanderplank harnessed underdominance, which occurs when hybrids of two strains are less fit than either, in an attempt to reduce the incidence of disease spread by tsetse flies (Vanderplank Citation1944, Citation1948). The approach was independently theorized by Serebrovskii and extended by C. F. Curtis using the specific example of chromosomal translocations (Serebrovskii Citation1940; Curtis Citation1968). Since then, scientists have proposed or actively attempted to harness numerous types of drive systems. Drawing from earlier works (Sinkins and Gould Citation2006; Marshall and Hay Citation2012; Alphey Citation2014; Burt Citation2014; Champer, Buchman, and Akbari Citation2016), these fall into two main classes based on their extent of geographic spread ().
Figure 1. Classification of gene drive systems by capacity to spread and reversibility using a subsequent drive system. Note that self-exhausting drive systems are all inherently sequence-reversible given enough time or the release of many wild-type organisms. A theoretical suppression method for Medea has been posited but is unlikely to be generalizable. Multi-locus assortment is not technically a drive system because it does not increase in frequency, but disperses to affect a larger fraction of the population.
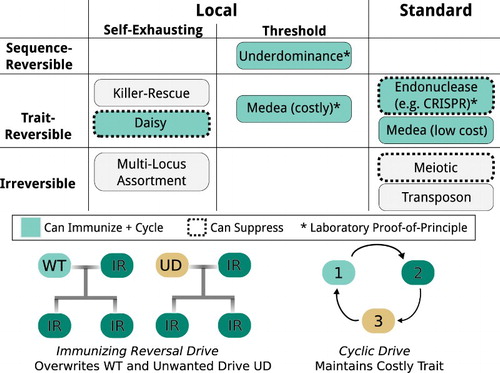
Gene drive classes
Standard drive systems may spread to all populations connected by gene flow. They are the only drive type capable of affecting very large populations and are the least costly to deploy. However, coordinating agreement among all potentially affected stakeholders may prove an insurmountable challenge in many cases. Examples include transposons that duplicate themselves within the genome (Carareto et al. Citation1997), meiotic drives that bias gametogenesis in their favor (Wood et al. Citation1977), costless Medea (Chen et al. Citation2007), and the many strategies proposed by Austin Burt that utilize homing endonucleases to cut chromosomes that do not encode them (Burt Citation2003).
Local drive systems can spread through regional populations, but cannot spread to all populations connected by gene flow. There are two sub-types of local drive systems.
Self-exhausting drive systems will initially spread to a larger fraction of the local population, then decrease in abundance without necessarily affecting every local organism connected by gene flow. They are suitable for geographic and temporally limited interventions, as well as expedient field tests to determine the ecological effects of more powerful drive systems. Examples include killer-rescue, which uses independent toxin and antitoxin genes to spread cargo genes associated with the antitoxin (Gould et al. Citation2008), and daisy drive systems, which separate the components of an endonuclease drive system into ‘daisy elements’ arranged in either a linear daisy-chain or parallel ‘daisyfield’ structure, and rely on the progressive loss of non-driving elements to limit the total spread of the cargo element (Noble et al. Citation2016; Min et al. Citation2017b). Daisy drives of two elements have been constructed as a ‘split drive’ confinement strategy (DiCarlo et al. Citation2015), but larger chains have not yet been demonstrated. Multi-locus assortment, while not technically a drive system, achieves a similar effect by releasing organisms carrying many independently assorting copies of a desired gene (Rasgon Citation2009), causing dispersal through the population without increasing in absolute frequency. Self-exhausting drive systems have previously been termed ‘self-limiting’, but the latter term has also been applied to sterile-male and other approaches that only ever decrease in frequency.
Threshold drive systems will spread to fixation if released above a threshold frequency but otherwise are rapidly driven extinct. They cannot be used when the wild population is large, but are well-suited to small islands or subpopulations with minimal gene flow. Examples include underdominance achieved through chromosomal translocations (Buchman et al. Citation2016) or toxin-antitoxin systems (Akbari et al. Citation2013; Reeves et al. Citation2014) as well as certain synthetic variants of the ‘Medea’ drive native to the Tribolium beetle (Beeman, Friesen, and Denell Citation1992; Wade and Beeman Citation1994; Chen et al. Citation2007; Marshall and Hay Citation2012; Akbari et al. Citation2014).
Finally, combination drive systems combine multiple types of drive. For example, a daisy quorum system combines a self-exhausting daisy drive with threshold-dependence to efficiently alter an entire local population without invading those nearby (Min et al. Citation2017a).
Drive systems within these classes vary in their degree of reversibility, ability to maintain costly alterations, capacity to suppress as well as alter populations, and whether engineered systems have been validated in the laboratory (Box 1).
Local drive systems exclusively affect local but not global populations.
Standard drive systems have the potential to affect every population of the target species.
Sequence-reversible systems can restore the original wild-type genome sequence. It cannot be distinguished from the natural state of a population.
Trait-reversible systems can restore the original phenotype, but not the exact DNA sequence. If done correctly, this is operationally indistinguishable from sequence reversibility and may allow for improved accounting, but some may value the natural state.
Irreversible systems spread alterations that cannot be undone using the same drive type.
Alteration drive systems directly change the DNA sequence and therefore the traits and behaviors of affected organisms. They can either add new transgenes or edit existing genes.
Suppression drive systems directly reduce the overall number of organisms in the population, potentially to extinction.
Immunizing drive systems spread through and recode the DNA of the wild-type population to block the spread of another drive system.
Cyclic drive systems can maintain costly traits by periodically overwriting broken versions.
Historical constraints
Attempts to harness gene drive systems have suffered from technological limitations, evolutionary instability, or both. Because genome editing was difficult, building and testing a theorized drive system in the laboratory required years of work in model organisms, let alone less genetically tractable pest species. Generating chromosomal translocations between two sequences of interest to create underdominance drive systems may have been theoretically possible, but was not feasible due to the dearth of molecular tools capable of efficiently targeting those sequences with sufficient precision in relevant species.
Engineering homing endonucleases to cut new sequences proved extremely difficult for both rational design and directed evolution (Thyme et al. Citation2014), forcing scientists to work with the rare natural enzymes that happened to target useful sequences (Windbichler, Papathanos, and Crisanti Citation2008).
A second problem concerned the evolutionary stability of gene drive systems, especially in the face of resistance. Natural meiotic drive systems are typically opposed by resistance elements and will not reach fixation in environments where they are present. Transposons did not spread as rapidly as anticipated when moved into new species (O’Brochta et al. Citation2003) and reliably shed inserted cargo genes (Carareto et al. Citation1997). Homing endonucleases, arguably even those that target repeated sequences (Burt Citation2003; Galizi et al. Citation2014), are vulnerable to drive-resistance alleles created when alternative repair pathways mutate the target site (Muscarella and Vogt Citation1993; Deredec, Burt, and Godfray Citation2008) or to recombination between repeated sequences within the drive construct (Simoni et al. Citation2014). Underdominance-based drive systems employing Medea-like toxins and antidotes can be vulnerable to loss of the toxin (Akbari et al. Citation2013).
The CRISPR era
The discovery and adaptation of CRISPR systems (Jinek et al. Citation2012; Cong et al. Citation2013; Mali et al. Citation2013) have dramatically accelerated gene drive research by making genome editing remarkably straightforward in many species. In organisms for which DNA can reliably be delivered into the germline, any type of synthetic drive system can in principle be inserted anywhere in the genome and subsequently optimized through iterative design-build-test cycles. This allows constructs to be optimized in a fraction of the time and may enable the creation of drive systems that were previously difficult to construct, such as chromosomal translocations between arbitrary genomic positions that could exhibit underdominance (Torres et al. Citation2014).
The basic mechanism of CRISPR genome editing involves supplying the CRISPR endonuclease, guide RNA(s) instructing it which sequences to cut, and templates encoding the edited sequences to be inserted. The cell then repairs the resulting double-strand break by incorporating the edited template sequence, a mechanism that is also utilized by naturally occurring homing nuclease gene drives. As a consequence, it is possible to build RNA-guided gene drive systems based on CRISPR endonucleases () (Esvelt et al. Citation2014).
Figure 2. The mechanism by which CRISPR gene drive systems distort inheritance. Adapted from Esvelt et al. (Citation2014).
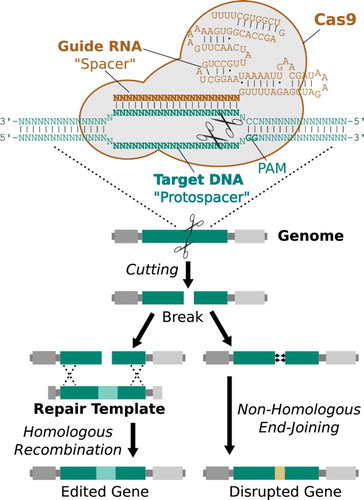
RNA-guided gene drive systems can cut any sequence in any sexually reproducing organism, enabling most types of alterations to be driven directly through wild populations singly or together. Like other endonuclease-based drive systems, they are self-sustaining and are consequently anticipated to spread to most populations of the target species. To date, CRISPR-based gene drive has been demonstrated in four different species (DiCarlo et al. Citation2015; Gantz and Bier Citation2015; Gantz et al. Citation2015; Hammond et al. Citation2016). The suppression mechanisms first outlined by Austin Burt are similarly accessible, one of which has already been demonstrated (Hammond et al. Citation2016).
Importantly, CRISPR-based gene drive systems can theoretically be rendered evolutionarily stable against drive-resistance alleles by using multiple guide RNAs to target many sequences within genes important for fitness (Esvelt et al. Citation2014). While not yet demonstrated in population genetics experiments, mathematical models suggest that this strategy should enable alterations to approach or even reach fixation (Noble et al. Citation2017) and ensure that suppression drives remain effective (Marshall et al. Citation2017).
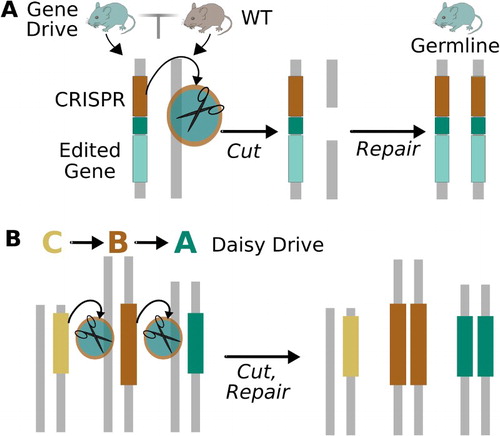
Standard CRISPR-based drive systems can be converted into equivalent local drive systems by separating the required components into a serially dependent ‘daisy-chain’ in which each element drives the next in the chain (). The resulting ‘daisy drive’ systems are constrained by the inability of the base element to drive, leading to the progressive loss of elements over generations. Adding links to the chain augments the spread of the final daisy element, permitting a small number of released organisms to alter much larger local populations (Noble et al. Citation2016). A parallel ‘daisyfield’ architecture produces similar results (Min et al. Citation2017b).
Figure 3. (a) A standard drive system based on CRISPR encodes all components necessary to drive itself. (b) A local ‘daisy drive’ system can be created by moving the components into different genetic loci such that each element drives the next in the chain. Because the basal element does not drive, the progressive loss of daisy elements over generations constrains the spread of the resulting ‘daisy drive’ system. The power of the daisy drive system can be tuned by changing the number of elements in the daisy-chain. Adapted from Noble et al. (Citation2016).
Applications
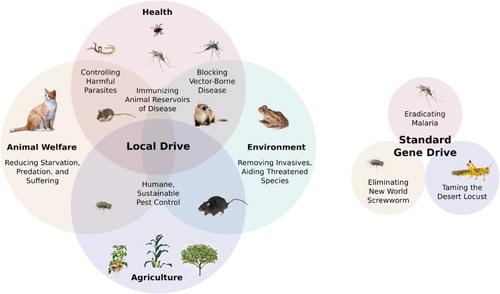
Gene drive systems could be used to address diverse ecological problems ranging from natural vector-borne and parasitic diseases to invasive species and pests transported by humans ().
Figure 4. Potential applications of local and standard gene drive systems. Because standard drive systems require agreement from every nation harboring the target species and likely cannot be tested in field trials without a substantial risk of unintended spread, there are only a handful of potentially feasible applications. Malaria is such a terrible scourge that any possible combination of unexpected effects would arguably be preferable to the disease itself. New World screwworm likely causes more animal suffering than any other wild species, is only present in South America, has already been eradicated from North America, and the effects of elimination can be tested locally using sterile insect technique. Desert locusts have been a hated agricultural plague in every culture afflicted since ancient times; they could be altered to remain solitary desert grasshoppers without threatening the existence of the species. Diplomatic and social barriers render other applications of standard drive systems highly unlikely; local drive systems do not necessarily face the problems.
Public health
The most pressing humanitarian problems are those in public health. Vector-borne diseases such as malaria, dengue, Zika, chikungunya, and others could be mitigated by altering the vector to block transmission or directly suppressing the vector. Parasites such the trematode worms that cause schistosomiasis could be directly targeted for eradication by suppression drives. In the developed world, Lyme and other diseases could be addressed through an ‘ecological vaccination’ approach in which heritable antibody-based immunity is spread through reservoir species such as white-footed mice (Tsao et al. Citation2004).
Agriculture
Modern agricultural practice relies heavily on a combination of herbicides, tilling, insecticides, or all three to control weeds and insect pests. As resistance to herbicides and pesticides evolves, farmers are left with less attractive options such as adopting entirely new compounds or redeploying older molecules that are less safe for humans or the environment. Most obviously, gene drive systems could directly reverse resistance in insects and certain weeds, giving safer compounds a new lease on life.
Alternatively, ‘sensitizing drives’ might confer vulnerability to new compounds, perhaps ones that are otherwise biologically inert and hence completely non-toxic to humans and the environment. This strategy would allow pests to be locally removed without affecting any other species or populations elsewhere.
Most elegantly, pests might be altered so that they no longer consume crops but otherwise perform their natural ecological functions. For example, advances in insect olfaction may eventually allow pests to be reprogramed to avoid human crops (Matthews et al. Citation2016). Another option might involve removing troublesome traits from pest species, such as the desert locust’s capacity to form gregarious swarms, thereby transforming them into harmless grasshoppers (Sugahara et al. Citation2015) without impacting the viability of the species (Chapuis et al. Citation2014). In some cases, cyclic drives would be necessary to maintain the deleterious trait in the target population (Esvelt et al. Citation2014).
Conservation
Species are continually invading new habitats as global travel and trade overcome geographic barriers. Some of these fail to establish, others are integrated relatively seamlessly into the existing ecosystem, but a handful becomes invasive. Excessive predation, direct competition, vectored diseases, and other indirect effects of invasive species are major ongoing causes of extinction worldwide, particularly on islands. For example, numerous native Hawaiian honeycreeper birds will likely go extinct absent intervention due to avian pox and avian malaria spread by introduced mosquitoes, the range of which is slowly expanding to encompass the last high-elevation bird habitats due to climate change (Atkinson and LaPointe Citation2009). Displacements of native organisms such as lake trout also have major negative impacts on industries such as fishing and tourism, while a number of invasive species such as the Asian citrus psyllid are also agricultural pests. Suppression drives may be capable of precisely eliminating invasive populations from non-native ranges in order to save native species such as the Hawaiian honeycreepers and help restore damaged ecosystems.
On the other hand, the removal of a species from an ecosystem can result in negative effects. The most dramatic changes are caused by the removal of keystone species, which results in large-scale changes to the stability of the entire ecosystem (Paine Citation1969). However, proposed targets of suppression drive systems such as mosquitoes and blood flukes are not keystone species. For the malaria vector Anopheles gambiae, a recent risk analysis workshop examining possible harms concluded that such risks exist, but are likely dwarfed by the impact of malaria (Roberts et al. Citation2017).
Why are major negative effects unlikely? First, it should be emphasized that the cost–benefit analysis for any proposed drive system will depend on the target species, ecosystem, and nature of the change in question: decisions must be made on a case-by-case basis. For malarial mosquitoes, a major reason is that there are so many different competing species likely to fill could fill the vacated niche of the targeted vector, causing predators to simply switch from one to the other. Moreover, no predators or pollinated plants are known to rely primarily on any one species of mosquito. Finally, the introduction of a population suppression drive systems would impose tremendous selection pressure favoring resistance mechanisms in the target population. That includes drive-resistant alleles (Marshall et al. Citation2017; Prowse et al. Citation2017), but also preferential inbreeding and asexual reproduction in capable species (Bull Citation2017; Drury et al. Citation2017). Given the size of wild populations and polymorphism rates, resistance to any single drive system, even one targeting many different sites, is bound to occur eventually. Just as diseases are never the cause of extinction on their own, it is implausible that any single suppression drive could cause the target species to go extinct – particularly when it could be rescued by any human capable of editing its genome. To reiterate: proposals to use suppression drive systems aim to suppress the target population, not to drive them extinct, and likely could not accomplish extinction by themselves without global consensus.
Technical limitations and candidate solutions
Attempts to realize gene drive applications face numerous technical challenges.
Sexual reproduction – Because distorting inheritance requires sexual reproduction events, gene drive systems will not function in viruses, bacteria, or archaea. Sexual organisms that primarily reproduce asexually (Bull Citation2017), self-fertilize, or even undergo high rates of inbreeding (Drury et al. Citation2017) are anticipated to be highly resistant, including yeast, some nematodes, and many plants.
Generation time – Being vertically transmitted, gene drive interventions targeting slowly reproducing species will be too slow to address problems in a human-relevant timeframe. As a rule of thumb, organisms with a generation time of more than 2 years are poor candidates.
Geographic Control – The control or elimination of invasive species or pests should ideally leave native populations unaffected. Because most suppression-capable drive systems are of the standard self-sustaining type, this presents a serious problem. For highly mobile species such as rats that are known to stow away on ships and planes, preventing spread to most populations may not be possible. Other species such as cane toads will not spread on their own, but could readily be deliberately transported by negligent individuals. If the drive system could offer economic benefits, the history of biocontrol introduction by individuals, even when illegal (O’Hara Citation2006), suggests deliberate unauthorized transport is extremely likely.
The only local drive system capable of direct population suppression is the daisy drive (Noble et al. Citation2016; Min et al. Citation2017b). If realized in relevant organisms, it could alleviate problems from both accidental and deliberate releases because the number of organisms released into unintended areas would be too small to affect more than a fraction of the local population.
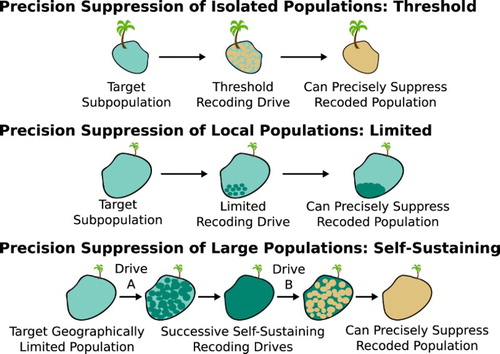
Another option is the ‘precision drive’ strategy, which involves using a suppression drive system to target a DNA sequence unique to the invasive population. Ideally, such sequences would exist already due to founder effects, but if not, they may be introduced using an alteration drive system () (Esvelt et al. Citation2014). When the invasive population is small, a threshold drive might be used for the initial recoding. If it is large, successive standard drive systems could obtain the same effect, albeit with a substantial risk of the first drive element spreading into the native population before replacement by the second. A combined daisy quorum drive system, if realized, would be ideally suited to this approach (Min et al. Citation2017a).
Figure 5. Precision drive systems can safely suppress small populations by first recoding them with a local drive, or in some cases by targeting natural mutations unique to a target population. Larger populations might rely on successive standard drives for recoding, but this risks the first recoding drive spreading into other populations before it is replaced by the second recoding drive. Adapted from Esvelt et al. (Citation2014).
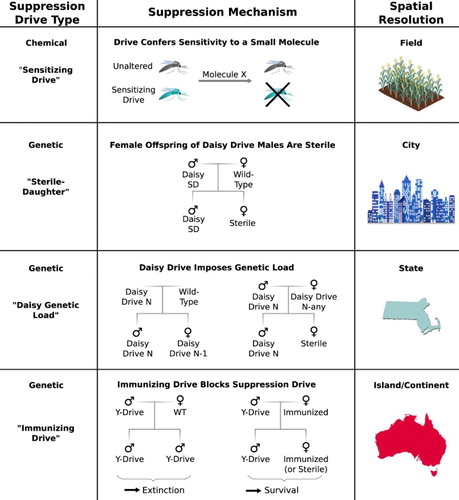
Drive systems could theoretically confer vulnerability to alternative methods of suppression () (Esvelt et al. Citation2014). Sensitizing drives could confer vulnerability to a small molecule, enabling very fine geographic control over suppression. In some species, alteration drives can be converted into unstable suppression drives by including male-determining genes as cargo. When the cargo mutates, suppression will cease, although the alteration drive would continue to spread. Finally, large native populations might be protected from suppression drives using an immunizing drive that recodes the relevant target sequence.
Figure 6. Methods of controlling the extent of population suppression using external triggers or barriers. Adapted from Esvelt et al. (Citation2014).
Evolutionary Stability – The suppression drive elements first described by Burt are inherently linked to the desired outcome: if the drive spreads at all, the population will be suppressed (Burt Citation2003). Because targeting multiple sites with CRISPR can theoretically block the creation of drive-resistant alleles, genetic load or sex-biasing suppression drives should remain stable over time, though unknown gene flow patterns could render local outcomes unpredictable (Deredec, Burt, and Godfray Citation2008; Beaghton, Beaghton, and Burt Citation2015). It is important to note that the target species may be rescued at any point if desired by releasing organisms engineered to carry drive-resistant alleles.
Alterations made by drive systems are not similarly stable because there is no direct selection pressure for the new sequence. Even without drive-resistant alleles or other mechanisms that directly block the drive element, the allele can mutate to restore the original trait, which will often be favored by natural selection. This can only be prevented by tightly linking the alteration to the drive mechanism, which is generally not possible and at best selects for the new trait until the drive reaches fixation. Including multiple copies of cargo genes or using an RNA-guided gene drive element to directly target potential unwanted sequences can buy time. In the long run, it is safe to assume that natural selection will eliminate all deleterious changes, even those enacted by standard or threshold drives.
Hence, establishing and maintaining costly changes may require creating successive generations of cyclic gene drive elements, each of which can overwrite the previous version and restore the functional element. CRISPR-based gene drives can readily accomplish this outcome (DiCarlo et al. Citation2015) as can underdominance and Medea (Akbari et al. Citation2014). However, the introduction threshold for underdominance and Medea become very high for large fitness costs.
Ease and versatility – An intervention is only useful if it can be realized. RNA-guided gene drive systems are self-inserting, well-understood from a molecular standpoint, have been demonstrated at least imperfectly in four species to date (DiCarlo et al. Citation2015; Gantz and Bier Citation2015; Gantz et al. Citation2015; Hammond et al. Citation2016), and rely on a mechanism known to function in essentially all organisms examined from a genome editing perspective (Esvelt et al. Citation2014). Building appropriate reversal, immunization, and cyclic drive systems are trivial once the initial drive has been optimized. Daisy drive systems are more complicated to build as they comprise multiple elements, but operate in an identical fashion (Noble et al. Citation2016). Synthetic Medea elements have been constructed in D. melanogaster using a method that may be generalizable to other species following characterization of early embryogenesis and oogenesis pathways (Chen et al. Citation2007; Akbari et al. Citation2014); subsequent drive systems will likely require an equivalent amount of effort. Toxin-based underdominance systems and killer-rescue systems are similar to Medea (Akbari et al. Citation2013; Reeves et al. Citation2014). Chromosomal translocations suitable for creating underdominance drive systems are presumably accessible in most CRISPR-amenable species, and have already been generated in Drosophila (Buchman et al. Citation2016). Multi-locus assortment is fairly straightforward to realize as it only requires cargo genes to be inserted at many locations in the target genome.
Side effects – Our knowledge of ecosystem-level species interactions is limited. Even when we know how to edit the genome to achieve the desired outcome, predicting its effects on other species is difficult. The best way to identify these effects is to release organisms carrying a local drive or no drive at all that are otherwise similar to the planned intervention and carefully study the consequences. Ideally, this should be done in all relevant habitats of the target species. Threshold gene drive systems can simply be released below the threshold. standard gene drive systems likely cannot be tested without probable spread to the entire population. However, a daisy drive system could be released to examine the likely effects of a standard CRISPR-based drive system operating by the same mechanism, while certain alterations could be tested by releasing organisms with multi-locus assortment or without any form of drive system.
Note that a handful of problems are so severe that it is highly implausible that unexpected ecological side effects could outweigh the benefit of addressing them; for example, no known human-caused ecological effect approaches the toll in human lives and suffering inflicted by malaria. This may limit the perceived need to experimentally identify likely side effects in the diverse ecosystems that might be affected by standard drive systems.
Social constraints and a path forwards
Gene drive systems are unique in their ability to impact many lives without requiring mass adoption in the marketplace. This capability raises troubling ethical issues.
It should go without saying that gene drive interventions must only be released with broad public support, yet throughout most of history, environmental impacts on other citizens were seldom factored into decision-making. Continuing this policy would be ethically and socially unacceptable in the modern era. Attempts to release gene drive systems without public involvement in the development process will likely cause future interventions to be rejected, even if the result is a net benefit to human life and the environment (Enserink Citation2008).
Moving forwards will require learning from past mistakes that were intrinsic to the current model of technology development. Much of the tension over genetically modified (GMOs) foods arose from (1) the dearth of obvious benefits to typical citizens, (2) the perceived absence of public notification or discussions prior to development, (3) a perceived failure to openly discuss safeguards or independent testing in advance of deployment, (4) a perceived failure to solicit and transparently address community concerns during development, and (5) public mistrust of for-profit corporations developing the technologies and performing safety tests. Gene drive researchers would do well to pursue the opposite approach in each case (Box 2) (Marris Citation2001; Durant and Legge Citation2006).
Develop interventions whose benefits are obvious to most citizens.
Invite public discussion of proposed projects before experiments begin.
Clearly detail safeguards that will be used.
Transparently address community concerns during the development cycle.
Ensure that early applications are community-directed and non-profit.
Following these guidelines will be considerably more straightforward when developing some types of gene drive systems rather than others.
First, it is easier to work closely with communities, raise awareness, and build public support when fewer people will be affected. What if one community seeks to rid itself of a disease, but the neighboring community does not consent? For a standard drive system, this is an irreconcilable conflict. For a local drive system, it may not be.
Second, people are more likely to support interventions that can be at least partly undone if something goes wrong. Full sequence reversibility is exclusive to certain threshold-dependent and combined drive systems and will be most attractive to people wary of permanently ‘contaminating’ nature. Such individuals may also support ‘cisgenic’ interventions that harness meiotic gene drives native to the species of interest, or costly self-exhausting drive systems that will be naturally purged from the population if not actively maintained through periodic releases. For the probable civic majority who do not place intrinsic value upon the original sequence, the faster spread of reversal drive systems based on CRISPR or possibly Medea-based systems could be a major advantage. Truly irreversible drive systems are unlikely to win support.
Third, interventions that are comparatively inexpensive to deploy may be more readily accomplished by non-profit organizations. Standard drive systems are far more cost-effective than most self-exhausting and threshold systems because the latter types require many more organisms to be released. However, daisy drive systems with many elements may require notably small release sizes.
Fourth, regulatory approval must be obtained from every country that would be affected by an eventual deployment. Because there is a strong argument that standard drive systems are likely to spread to most populations of the target species, they are likely to be limited to a handful of applications targeting relatively geographically confined species.
Together, these analyses suggest that self-exhausting and/or threshold drive systems are likely to be required for almost all applications. Standard drive systems may be required to address serious problems involving very large wild populations, most notably malaria and the New World screwworm.
Moral hazards and technical safeguards
Most biologists can safely assume that their experiments will only impact people physically present in the laboratory. Transgenesis experiments in organisms such as fruit flies can reasonably be assumed harmless because natural selection will reliably eliminate any escaped mutant flies. With the advent of standard gene drive systems, this situation inverts: the default expectation is that escaped organisms will lead to the drive system spreading to at least some fraction of the wild population.
Scientists have a moral obligation to ensure that laboratory accidents cannot directly affect the outside world. Given the media spotlight that would doubtless focus on any unauthorized gene drive release, even researchers who are skeptical that gene drive systems will spread so readily should carefully consider the consequences of an error. At stake are not just personal and scientific integrity, but also public trust in science, the future prospects of gene drive interventions, and all of the lives and species that might otherwise be saved.
Any accidental gene drive release would conclusively demonstrate that at least some scientists cannot be trusted to work with such a powerful technology. Once lost, trust is difficult to regain (Cvetkovich et al. Citation2002; Renn and Levine Citation1991; Black and Rappuoli Citation2010). Whether the drive causes any ecological damage or spreads to more than a fraction of the entire wild population is unlikely to matter in the court of public opinion. Regardless of the cause, prominent mistakes causing loss of public trust can be devastating; the tragic death of Jesse Gelsinger set back the field of gene therapy by over a decade (Wilson Citation2009). Because gene drive releases will require broad support – including from members of the public already suspicious of genetically modified organisms, as well as those who view nature with religious reverence – the political and public relations fallout of an accidental gene drive release would likely be far worse than the Gelsinger tragedy. Given that community-supported gene drive interventions could potentially control diseases that cost thousands of lives every day, there is a tremendous moral imperative to avoid mistakes that could delay deployment.
Sources of skepticism
Many scientists worry that social pressure or additional regulations mandating safeguards will delay life-saving research (Spradling Citation2015). While the concerns of these researchers may be well-founded, they assume that benevolent technologies will always be adopted, or at least that the scientific community will be the final arbiter. Today, even vaccines face widespread objections, and that controversy is minor compared to the battle over GMO foods: according to a recent poll, 88% of scientists believe that GMO foods are safe to eat, but only 37% of US adults agree. This is currently the single largest opinion gap between scientists and society (Funk and Rainie Citation2015). If not developed in a different manner, gene drive technology is likely to be at least as divisive – and because there is no opportunity for citizens to choose whether or not to purchase gene drive products of their own initiative, this level of skepticism will preclude ethical deployment.
Other researchers may be justifiably skeptical that synthetic constructs based on current standard gene drive technologies will in fact spread in the wild. If we are far away from success, caution at this moment would arguably waste time and effort. A number of species possess intrinsic defenses against natural gene drive systems such as transposons; these may also block synthetic drive systems. But just because defenses exist does not mean that they will be present in species of interest, or that anti-transposon defenses in multicellular eukaryotes will be effective against CRISPR drive systems that their ancestors never encountered. Even effective defenses could conceivably be directly targeted and destroyed by a CRISPR system upon fertilization. Moreover, any defense capable of completely blocking spread in the wild should be observable in the laboratory when testing the drive system against multiple strains. To date, no such phenomenon has been observed.
Of the published CRISPR gene drive systems, the gene drive systems constructed in Anopheles were handicapped by sex-specific activity due to the use of a promoter that resulted in the maternal deposition of active Cas9 into the oocyte (Gantz et al. Citation2015; Hammond et al. Citation2016). Even so, the Anopheles stephensi alteration drive system still exhibited drive strong enough to spread given a low fitness cost. A combination of using a different promoter and decreasing Cas9 stability (Galizi et al. Citation2014) could address the deposition problem. More relevantly, published CRISPR drive systems in D. melanogaster exhibited drive likely sufficient for the spread in the wild (Gantz and Bier Citation2015; Champer et al. Citation2017) if the fitness cost of the cDNA or promoter disrupted is not too high for the drive efficiency (Drapeau et al. Citation2006). Other drive types might also function in the wild; in particular, the synthetic Medea element created almost a decade ago could conceivably spread if migration rates were high enough to overcome the low level of threshold-dependence imposed by the fitness cost (Chen et al. Citation2007).
As transgenesis methods for non-model organisms improve, the number of accessible species will increase. In short, many researchers currently or soon will have the ability to unilaterally alter the shared environment. Scientists must remain mindful that great power entails equally great responsibility, and take precautions accordingly.
Safeguards for laboratory studies
Confinement strategies applicable to CRISPR gene drive systems have already been detailed (Benedict et al. Citation2008; Esvelt et al. Citation2014), publicized (Oye et al. Citation2014; Akbari et al. Citation2015), and demonstrated (DiCarlo et al. Citation2015). Many require little or no effort to implement; some would also apply to Medea drive systems. These safeguards fall into two major classes.
Extrinsic confinement
Barrier confinement prevents gene drive organisms from escaping the laboratory (). Barriers are the traditional first line of defense and can be very effective when adapted to the idiosyncrasies of each organism. Laboratories considering barrier confinement of gene drive organisms, especially highly mobile species such as flying insects, should refer to the expertise developed by ecologists employing biocontrol. However, barriers are intrinsically vulnerable to human error, as has been extensively documented for work with dangerous pathogens, and to deliberate release by unauthorized humans. With a standard drive system, one mistake is likely to suffice (Marshall Citation2009).
Figure 7. Extrinsic confinement strategies aim to prevent organisms carrying gene drive constructs from mating with wild counterparts. They are vulnerable to human error and to deliberate interference.

Ecological confinement is accomplished by performing experiments in geographic areas lacking populations of the organism in question. Ideally, the organism will not be able to survive outdoors: tropical Anopheles malarial mosquitoes would not survive winters in Boston or London. While less vulnerable to human error than barrier confinement, researchers traveling to the native habitat could also inadvertently transport organisms in their clothing or luggage. Refraining from visiting the laboratory for several days prior to such a trip may be advisable. Ecological confinement is also vulnerable to deliberate transport by unauthorized individuals.
Intrinsic confinement
Reproductive confinement involves the use of laboratory organisms that cannot reproduce with wild counterparts and consequently cannot pass on the gene drive system (). For example, some laboratory strains of D. melanogaster have a ‘compound autosome’ formed by conjoining both copies of a large autosome centromere (Fitz-Earle, Holm, and Suzuki Citation1973; Cantelo and Childress Citation1974). These strains are fertile when crossed amongst themselves, but are sterile when outcrossed to any normal or wild-type strain because all progeny are monosomic or trisomic and die early in development. However, some of these strains are sickly and these types of lines are not available for most organisms.
Figure 8. Intrinsic confinement strategies are intended to prevent population alteration even if organisms carrying gene drive constructs mate with wild counterparts. They are inherent to the organism or the gene drive system and once created are not vulnerable to human error or deliberate interference.
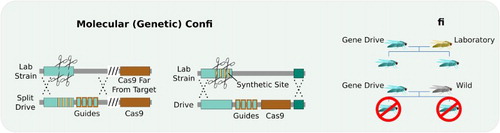
Molecular confinement, also termed genetic confinement, ensures that the gene drive system is constructed such that it cannot sustainably bias inheritance in a wild population. There are two orthogonal approaches.
Split drive systems separate crucial components such that only one of them can exhibit drive (Esvelt et al. Citation2014). A drive system encoding guide RNAs but no nuclease gene will spread readily in transgenic organisms that already express Cas9 from an unlinked locus, but cannot bias inheritance without it. Should organisms accidentally escape the laboratory, the population frequency of the Cas9 gene will be determined by normal Mendelian dynamics and consequently limit the spread of the sgRNA cassette to a rate no different than any other genetically modified organism. In yeast, sgRNA-only systems are copied as efficiently as Cas9 + sgRNA systems (DiCarlo et al. Citation2015). An important caveat is that recombination events could theoretically transform a split drive into an autonomous drive system. This is also true for daisy drive systems (Noble et al. Citation2016). Analogously, the split drive version of a Medea system is equivalent to local drive killer-rescue systems.
Synthetic site targeting involves constructing gene drive systems that exclusively cut synthetic sequences not found in wild populations (Esvelt et al. Citation2014). They only exhibit drive in transgenic laboratory populations carrying the synthetic sequence (DiCarlo et al. Citation2015). Because a large number of mutations within the guide RNAs would be required to escape, synthetic site targeting is almost certainly the most robust confinement strategy available. It is also applicable to synthetic Medea constructs and other drive systems based on RNAi toxins.
Awareness and consensus
Earlier discussions among researchers concerned with gene drive and vector-borne disease led to guidelines for field trials of standard gene drive systems (Benedict et al. Citation2008; WHO Citation2014). Individual groups additionally called for laboratory-level safeguards in advance of experiments involving CRISPR gene drive (Esvelt et al. Citation2014; Oye et al. Citation2014).
However, safeguards and guidelines are only useful if scientists are aware of them and proceed accordingly. This reality was underscored by the construction of the first CRISPR-based gene drive construct in fruit flies (Gantz and Bier Citation2015). These experiments were performed for reasons unrelated to population editing by scientists who at the time were not aware of earlier research on gene drive systems and applicable safeguards other than traditional barrier confinement. As a highly mobile species famous for escaping the laboratory and now endemic worldwide, the fruit fly arguably poses the greatest risk of accidental release. Fortunately, both reproductive and molecular confinement are accessible options in fruit flies; indeed, basic split drive systems are easier to construct than are standard CRISPR gene drive constructs thanks to the ready availability of numerous Cas9-expressing strains (Port et al. Citation2014).
To address the problem, scientists convened a working group including many other researchers in the fruit fly and gene drive fields to establish consensus recommendations. The result was a highly publicized call for laboratories working with standard drive systems to use multiple stringent confinement strategies whenever possible (Akbari et al. Citation2015). These are likely to be superseded by formal national and international guidelines.
Safeguards for field trials
Standard gene drive systems that are candidates for eventual release cannot employ intrinsic confinement. Ecological confinement should consequently be used whenever possible (Benedict et al. Citation2008). Caged field trials and remote island trials are theoretically options, but potential targets such as the invasive fruit fly pest D. suzukii (Li and Scott Citation2015) are just as prone to escaping cages as the model fruit fly D. melanogaster, and remote island experiments may need to be subjected to military-level quarantine to prevent deliberate unauthorized transport.
The only way to ensure that an accidental release does not occur is to avoid releasing organisms encoding standard gene drive constructs expected to function in the wild. As discussed above, releasing organisms encoding self-exhausting or non-driving systems with similar effects may be the optimal strategy; these could be followed by standard drive systems specific to the alteration.
As a final precaution, drive systems intended for release could be created in tandem with immunizing reversal drives capable of overwriting the relevant sequence change and blocking further spread through the as-yet unaffected population (Esvelt et al. Citation2014). Building an immunizing reversal drive system is no more difficult than constructing the first drive system and in many cases will be easier. Notably, immunizing reversal is projected to be possible using local as well as standard drive systems (Min et al. Citation2017a). Ensuring that these countermeasures are available in advance will help to mitigate ecological and social damage caused by accidental releases or unexpected side effects. Any efforts to develop interventions that fail to take such precautions may be viewed as less responsible. Future advances capable of perfectly restoring the population to its original genetic state could be transformative for both safety and public acceptance.
Potential for misuse
In an ideal world, synthetic gene drive systems would only ever be used to benefit humanity and the environment. Regrettably, the possibility of misuse does exist. The extent to which CRISPR gene drive could be used as a weapon was carefully considered prior to public disclosure of the technology. The risk is far less severe than many accounts in the popular press would have it, and also remains years away. However, minimizing it will require active monitoring.
Because social unease at the prospect of misused gene drive technology could impede public support for development, it is worth outlining the reasons why attempts to use gene drive as a weapon would be comparatively ineffective. Three factors combine to strongly favor defenses against misuse.
Universal detection: High-throughput sequencing can unfailingly detect a gene drive system in any given organism and perhaps even in environmental metagenomic samples. The arrangement of the requisite genes cannot be hidden and does not occur in nature. No technology is falling as rapidly in cost as genome sequencing, so detection should only become easier with time.
Universal overwriting: Given the diverse protospacer-adjacent motifs that can be targeted by different Type II and Type V CRISPR systems, it is not currently possible to build a gene drive that cannot be overwritten by another drive system, nor is this likely to change in the foreseeable future. Once detected, any drive element can be overwritten by an immunizing reversal drive constructed for the purpose. Especially if it uses the same nuclease and expression conditions, the countermeasure will exhibit drive as efficient as the original and will overwrite both the unwanted element and the wild-type version.
Slow spread: It is difficult to release large numbers of organisms without attracting attention; defense agencies are under no such constraint. Whereas the frequency of a gene drive element can at most double in any given generation, most fast-reproducing organisms are r-selected and have dozens to thousands of offspring. This enables large numbers of organisms carrying an immunizing reversal drive to be bred and continually released in a geographic pattern designed to overwrite and swiftly erase the unwanted element soon after detection.
To be clear, misused gene drive systems could cause harm before being detected and countered, but that harm would be comparatively minor as long as monitoring is adequate - especially relative to alternative approaches available to anyone with the resources and technical knowledge required to attempt the assault. However, the outlined defense strategy will require agencies to monitor populations and environments considered at-risk through environmental sequencing, which will cost resources. Ideally, the resulting data will be used to benefit science and the environment as well as biosafety.
A step towards scientific reform
Most science and technology development currently takes place behind closed doors. This system is inefficient: duplication of effort and the failure to publish experiments that did not succeed guarantee wasted time and money. It decreases public trust: a common complaint to regulatory agencies concerns the lack of access to raw data. In some cases it is of dubious morality: mistakes during research on gene drive systems, geoengineering, and potential pandemic pathogens could directly impact people beyond the laboratory or field trial site. In some cases it may even be dangerous, because few scientists can adequately assess the broader consequences of their work when combined with other powerful technologies of which they are unaware.
Modern science amounts to a collaborative venture in which all participants have agreed to keep the lights off during development. It is questionable whether there are any advantages to the current system; while competition could spur heightened effort and creativity, it is unclear why the same benefits could not be obtained in an open system. More, the pressure-cooker atmosphere of modern science is harmful to researcher morale and well-being, damages work–life balance, and arguably encourages unethical behavior (Petersen et al. Citation2012; Couzin-Frankel Citation2014; Fang and Casadevall Citation2015).
No one starting from scratch would rationally design the current scientific enterprise (Tijdink et al. Citation2016). Yet systemic incentives block reform by punishing researchers who disclose their research plans before experiments begin and share results before publication. Those willing to share are seldom – if ever – rewarded for their ideas, but always make themselves vulnerable to being ‘scooped’ by better-funded or better-staffed laboratories. The potential advantages in the form of increased collaborations evidently do not make up for the perceived risks.
The nascent field of gene drive research has an opportunity to take a different path due to unique ethical and practical questions. Can scientists justify performing experiments behind closed doors when a mistake could directly impact people outside the laboratory? Is it wise to permit continued closed-door studies when an accidental release by an unknowing researcher could perhaps irreparably damage public trust and the efforts of everyone else in the field? Should communities trust scientists who insist on doing their work in secret? An open, community-responsive approach is far more likely to win public backing, without which gene drive projects are unlikely to reach fruition. There are legitimate differences of opinion on the ethics of developing and releasing life-saving interventions with shared impacts, but many fewer disagreements when it comes to whether shared-impact science would – in an ideal world – be done in the open. And if one field successfully implements a new model and changes the incentive structure accordingly, it will be considerably easier for that model to spread.
In short, the field of gene drive may have an opportunity to sow the seeds of scientific reform by taking an early step towards pre-registered research and responsible innovation (Esvelt Citation2016, Citation2017). Discussions between many researchers in the field have revealed that all would prefer to work in the open, but many are understandably reluctant unless everyone in the field agrees to follow the new model. More, such agreements would not necessarily preclude outsiders from conducting gene drive research in secret. Our task would be considerably easier with outside assistance: we need to change the incentive system, which hinges on recognition and funding.
To encourage early sharing, journals could reward researchers who pre-register their plans in advance of performing experiments by offering appropriate recognition once those ideas are experimentally validated. For example, journals might create a new type of Perspective reserved for those who first disclosed key aspects of the proposed and now experimentally validated the approach, even if they are not the first to validate it. Declining to publish the results of gene drive experiments that were not pre-registered could incentivize researchers to abide by new community norms. Similarly, funders or journals could make public disclosure of grant proposals and periodic updates a requirement to receive support and publish the outcome. Marshaling such a coordinated effort by the scientific community, journals, and funders would be exceedingly difficult in any other field. For gene drive, the questionable ethics of conducting experiments intended to alter the shared environment behind closed doors may make it possible.
Discussion
Scientists have long sought a way to alter wild populations in order to combat the vector-borne disease. CRISPR genome editing has brought many hypothesized gene drive systems within reach. By directly altering the genomes of wild populations, gene drive systems offer far more elegant solutions to ecological problems than are accessible to traditional physical, chemical, or engineering approaches. With the notable exception of malaria, diplomatic challenges are likely to preclude almost all applications of standard gene drive systems anticipated to spread to most populations of the target species. Hence, the development of local drive systems offering local community control is likely to be required. All such applications pose ethical challenges quite different from earlier technologies due to the potential for unilateral alteration of the shared environment.
The recent realization of predicted CRISPR-based gene drive systems in multiple species highlights the urgent need for scientists to understand the technical and social constraints complicating responsible development. Researchers are obligated to perform experiments with caution to prevent any accidental release that might affect non-consenting citizens outside the laboratory, an event that would certainly devastate public trust and set back gene drive interventions by years. At the same time, they face a moral obligation to accelerate development in order to reduce human suffering and prevent extinctions.
Successfully balancing these conflicting demands will require a new approach to science and technology development that emphasizes openness, responsiveness, and community guidance of interventions designed to impact the shared environment. Awareness of the issues, the availability and need for safeguards, and the strengths and weaknesses of different gene drive systems can help scientists walk this delicate tightrope. We are, at long last, learning to speak the language of nature. With sufficient cooperation and humility, we might even use it wisely.
Acknowledgements
We thank J. Delborne, A. Snow, L. Carter, and L. Alphey for their helpful comments.
Notes on contributors
John Min is a graduate student at the MIT Media Lab and the Department of Genetics at Harvard Medical School. He was a leader in developing the concept of the daisy drive and is pioneering efforts to study gene drive safety and evolutionary dynamics in nematode worms.
Andrea L. Smidler is a graduate student in the Church and Catteruccia laboratories at Harvard Medical School and Harvard School of Public Health. She was among the first to realize the potential of CRISPR systems to build gene drive systems and is currently constructing the first evolutionarily stable drive system in a mosquito.
Devora Najjar is a graduate student at the MIT Media Lab focusing on ethically-minded bioengineering projects. Currently working to develop the first mammalian daisy drive systems, she has a strong interest in the implications for biotechnology policy, biosecurity, and public discussions.
Kevin M. Esvelt is an assistant professor and leader of the Sculpting Evolution Group at the MIT Media Laboratory. He was among the first to describe the promise and peril of RNA-guided gene drive systems, including daisy drives, and has been leading the effort to promote early-stage transparency, safeguards, and public guidance of the technology.
ORCID
Andrea L. Smidler http://orcid.org/0000-0002-3281-1161
Devora Najjar http://orcid.org/0000-0001-9048-3592
Kevin M. Esvelt http://orcid.org/0000-0001-8797-3945
Additional information
Funding
References
- Akbari, Omar S., Hugo J. Bellen, Ethan Bier, Simon L. Bullock, Austin Burt, George M. Church, Kevin R. Cook, et al. 2015. “BIOSAFETY. Safeguarding Gene Drive Experiments in the Laboratory.” Science (New York, N.Y.) 349 (6251): 927–929. doi:10.1126/science.aac7932.
- Akbari, Omar S., Chun-Hong Chen, John M. Marshall, Haixia Huang, Igor Antoshechkin, and Bruce A. Hay. 2014. “Novel Synthetic Medea Selfish Genetic Elements Drive Population Replacement in Drosophila; a Theoretical Exploration of Medea-Dependent Population Suppression.” ACS Synthetic Biology 3 (12): 915–928. doi:10.1021/sb300079h.
- Akbari, Omar S., Kelly D. Matzen, John M. Marshall, Haixia Huang, Catherine M. Ward, and Bruce A. Hay. 2013. “A Synthetic Gene Drive System for Local, Reversible Modification and Suppression of Insect Populations.” Current Biology: CB 23 (8): 671–677. doi:10.1016/j.cub.2013.02.059.
- Alphey, Luke. 2014. “Genetic Control of Mosquitoes.” Annual Review of Entomology 59: 205–224. doi:10.1146/annurev-ento-011613-162002.
- Atkinson, Carter T., and Dennis A. LaPointe. 2009. “Introduced Avian Diseases, Climate Change, and the Future of Hawaiian Honeycreepers.” Journal of Avian Medicine and Surgery 23 (1): 53–63. doi:10.1647/2008-059.1.
- Beaghton, Andrea, Pantelis John Beaghton, and Austin Burt. 2015. “Gene Drive Through a Landscape: Reaction-Diffusion Models of Population Suppression and Elimination by a Sex Ratio Distorter.” Theoretical Population Biology 108 (December): 51–69. doi:10.1016/j.tpb.2015.11.005.
- Beeman, R. W., K. S. Friesen, and R. E. Denell. 1992. “Maternal-Effect Selfish Genes in Flour Beetles.” Science (New York, N.Y.) 256 (5053): 89–92. doi: 10.1126/science.1566060
- Benedict, M., P. D’Abbs, S. Dobson, M. Gottlieb, L. Harrington, S. Higgs, A. James, et al. 2008. “Guidance for Contained Field Trials of Vector Mosquitoes Engineered to Contain a Gene Drive System: Recommendations of a Scientific Working Group.” Vector Borne and Zoonotic Diseases (Larchmont, N.Y.) 8 (2): 127–166. doi:10.1089/vbz.2007.0273.
- Black, Steven, and Rino Rappuoli. 2010. “A Crisis of Public Confidence in Vaccines.” Science Translational Medicine 2 (61): 61mr1. doi:10.1126/scitranslmed.3001738.
- Buchman, Anna B., Tobin Ivy, John M. Marshall, Omar Akbari, and Bruce A. Hay. 2016. “Engineered Reciprocal Chromosome Translocations Drive High Threshold, Reversible Population Replacement in Drosophila.” BioRxiv, 088393. doi:10.1101/088393.
- Bull, James J. 2017. “OUP: Lethal Gene Drive Selects Inbreeding.” Evolution, Medicine, and Public Health 2017 (1): 1–16. doi:10.1093/emph/eow030.
- Burt, Austin. 2003. “Site-specific Selfish Genes as Tools for the Control and Genetic Engineering of Natural Populations.” Proceedings of the Royal Society B: Biological Sciences 270 (1518): 921–928. doi:10.1098/rspb.2002.2319.
- Burt, Austin. 2014. “Heritable Strategies for Controlling Insect Vectors of Disease.” Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 369 (1645): 20130432. doi:10.1098/rstb.2013.0432.
- Burt, Austin, and Robert Trivers. 2008. Genes in Conflict: The Biology of Selfish Genetic Elements. Cambridge, MA: Harvard University Press.
- Cantelo, W. W., and D. Childress. 1974. “Laboratory and Field Studies with a Compound Chromosome Strain of Drosophila Melanogaster.” Theoretical and Applied Genetics 45 (1): 1–6. doi:10.1007/BF00281167.
- Carareto, C. M. A., W. Kim, M. F. Wojciechowski, P. O’Grady, A. V. Prokchorova, J. C. Silva, and M. G. Kidwell. 1997. “Testing Transposable Elements as Genetic Drive Mechanisms Using Drosophila P Element Constructs as a Model System.” Genetica 101 (1): 13–33. doi: 10.1023/A:1018339603370
- Champer, Jackson, Anna Buchman, and Omar S. Akbari. 2016. “Cheating Evolution: Engineering Gene Drives to Manipulate the Fate of Wild Populations.” Nature Reviews Genetics 17 (3): 146–159. doi:10.1038/nrg.2015.34.
- Champer, Jackson, Riona Reeves, Suh Yeon Oh, Chen Liu, Jingxian Liu, Andrew G. Clark, Philipp W. Messer, and Harmit S. Malik. 2017. “Novel CRISPR/Cas9 Gene Drive Constructs Reveal Insights into Mechanisms of Resistance Allele Formation and Drive Efficiency in Genetically Diverse Populations.” PLOS Genetics 13 (7): e1006796. doi:10.1371/journal.pgen.1006796.
- Chapuis, Marie-Pierre, Christophe Plantamp, Laurence Blondin, Christine Pagès, Jean-Michel Vassal, and Michel Lecoq. 2014. “Demographic Processes Shaping Genetic Variation of the Solitarious Phase of the Desert Locust.” Molecular Ecology 23 (7): 1749–1763. doi:10.1111/mec.12687.
- Chen, Chun-Hong, Haixia Huang, Catherine M. Ward, Jessica T. Su, Lorian V. Schaeffer, Ming Guo, and Bruce A. Hay. 2007. “A Synthetic Maternal-Effect Selfish Genetic Element Drives Population Replacement in Drosophila.” Science (New York, N.Y.) 316 (5824): 597–600. doi:10.1126/science.1138595 doi: 10.1126/science.1138595
- Cong, Le, F. Ann Ran, David Cox, Shuailiang Lin, Robert Barretto, Naomi Habib, Patrick D. Hsu, et al. 2013. “Multiplex Genome Engineering Using CRISPR/Cas Systems.” Science (New York, N.Y.) 339 (6121): 819–823. doi:10.1126/science.1231143.
- Couzin-Frankel, Jennifer. 2014. “Chasing the Money.” Science 344 (6179): 24–25. doi:10.1126/science.344.6179.24.
- Curtis, C. F. 1968. “Possible Use of Translocations to Fix Desirable Genes in Insect Pest Populations.” Nature 218 (5139): 368–369. doi: 10.1038/218368a0
- Cvetkovich, George, Michael Siegrist, Rachel Murray, and Sarah Tragesser. 2002. “New Information and Social Trust: Asymmetry and Perseverance of Attributions about Hazard Managers.” Risk Analysis: An Official Publication of the Society for Risk Analysis 22 (2): 359–367. doi: 10.1111/0272-4332.00030
- Deredec, Anne, Austin Burt, and H. C. J. Godfray. 2008. “The Population Genetics of Using Homing Endonuclease Genes in Vector and Pest Management.” Genetics 179 (4): 2013–2026. doi:10.1534/genetics.108.089037.
- DiCarlo, James E., Alejandro Chavez, Sven L. Dietz, Kevin M. Esvelt, and George M. Church. 2015. “Safeguarding CRISPR-Cas9 Gene Drives in Yeast.” Nature Biotechnology 33 (12): 1250–1255. doi:10.1038/nbt.3412.
- Drapeau, Mark David, Shawn A. Cyran, Michaela M. Viering, Pamela K. Geyer, and Anthony D. Long. 2006. “A Cis-Regulatory Sequence Within the Yellow Locus of Drosophila Melanogaster Required for Normal Male Mating Success.” Genetics 172 (2): 1009–1030. doi:10.1534/genetics.105.045666.
- Drury, Douglas W., Amy L. Dapper, Dylan J. Siniard, Gabriel E. Zentner, and Michael J. Wade. 2017. “CRISPR/Cas9 Gene Drives in Genetically Variable and Nonrandomly Mating Wild Populations.” Science Advances 3 (5): e1601910. doi:10.1126/sciadv.1601910.
- Durant, Robert F., and Jerome S. Legge. 2006. “‘Wicked Problems,’ Public Policy, and Administrative Theory Lessons From the GM Food Regulatory Arena.” Administration & Society 38 (3): 309–334. doi:10.1177/0095399706289713.
- Enserink, Martin. 2008. “Tough Lessons from Golden Rice.” Science (New York, N.Y.) 320 (5875): 468–471. doi:10.1126/science.320.5875.468.
- Esvelt, Kevin. 2016. “Gene Editing Can Drive Science to Openness.” Nature 534 (7606): 153–153. doi:10.1038/534153a.
- Esvelt, Kevin M. 2017. “Precaution: Open Gene Drive Research.” Science (New York, N.Y.) 355 (6325): 589–590. doi:10.1126/science.aal5325.
- Esvelt, Kevin M., Andrea L. Smidler, Flaminia Catteruccia, and George M. Church. 2014. “Concerning RNA-Guided Gene Drives for the Alteration of Wild Populations.” ELife, e03401. doi:10.7554/eLife.03401.
- Fang, Ferric C., and Arturo Casadevall. 2015. “Competitive Science: Is Competition Ruining Science?” Infection and Immunity 83 (4): 1229–1233. doi: 10.1128/IAI.02939-14
- Fitz-Earle, M., D. G. Holm, and D. T. Suzuki. 1973. “Genetic Control of Insect Populations: I. Cage Studies of Chromosome Replacement by Compound Autosomes in Drosophila Melanogaster.” Genetics 74 (3): 461–475.
- Funk, Cary, and Lee Rainie. 2015. “Public and Scientists’ Views on Science and Society.” Pew Research Center: Internet, Science & Tech. January 29. http://www.pewinternet.org/2015/01/29/public-and-scientists-views-on-science-and-society/.
- Galizi, Roberto, Lindsey A. Doyle, Miriam Menichelli, Federica Bernardini, Anne Deredec, Austin Burt, Barry L. Stoddard, Nikolai Windbichler, and Andrea Crisanti. 2014. “A Synthetic Sex Ratio Distortion System for the Control of the Human Malaria Mosquito.” Nature Communications 5 (June), doi:10.1038/ncomms4977.
- Gantz, Valentino M., and Ethan Bier. 2015. “Genome Editing. The Mutagenic Chain Reaction: A Method for Converting Heterozygous to Homozygous Mutations.” Science (New York, N.Y.) 348 (6233): 442–444. doi:10.1126/science.aaa5945.
- Gantz, Valentino M., Nijole Jasinskiene, Olga Tatarenkova, Aniko Fazekas, Vanessa M. Macias, Ethan Bier, and Anthony A. James. 2015. “Highly Efficient Cas9-Mediated Gene Drive for Population Modification of the Malaria Vector Mosquito Anopheles Stephensi.” Proceedings of the National Academy of Sciences 112 (49): E6736–E6743. doi:10.1073/pnas.1521077112.
- Gould, Fred. 2008. “Broadening the Application of Evolutionarily Based Genetic Pest Management.” Evolution; International Journal of Organic Evolution 62 (2): 500–510. doi:10.1111/j.1558-5646.2007.00298.x.
- Gould, Fred, Yunxin Huang, Mathieu Legros, and Alun L. Lloyd. 2008. “A Killer-Rescue System for Self-Limiting Gene Drive of Anti-Pathogen Constructs.” Proceedings. Biological Sciences/The Royal Society 275 (1653): 2823–2829. doi:10.1098/rspb.2008.0846.
- Hammond, Andrew, Roberto Galizi, Kyros Kyrou, Alekos Simoni, Carla Siniscalchi, Dimitris Katsanos, Matthew Gribble, et al. 2016. “A CRISPR-Cas9 Gene Drive System Targeting Female Reproduction in the Malaria Mosquito Vector Anopheles Gambiae.” Nature Biotechnology 34 (1): 78–83. doi:10.1038/nbt.3439.
- Jinek, Martin, Krzysztof Chylinski, Ines Fonfara, Michael Hauer, Jennifer A. Doudna, and Emmanuelle Charpentier. 2012. “A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity.” Science (New York, N.Y.) 337 (6096): 816–821. doi:10.1126/science.1225829.
- Li, Fang, and Maxwell J. Scott. 2015. “CRISPR/Cas9-Mediated Mutagenesis of the White and Sex Lethal Loci in the Invasive Pest, Drosophila Suzukii.” Biochemical and Biophysical Research Communications, doi:10.1016/j.bbrc.2015.12.081.
- Mali, Prashant, Luhan Yang, Kevin M. Esvelt, John Aach, Marc Guell, James E. DiCarlo, Julie E. Norville, and George M. Church. 2013. “RNA-Guided Human Genome Engineering via Cas9.” Science (New York, N.Y.) 339 (6121): 823–826. doi:10.1126/science.1232033.
- Marris, Claire. 2001. “Public Views on GMOs: Deconstructing the Myths.” EMBO Reports 2 (7): 545–548. doi:10.1093/embo-reports/kve142.
- Marshall, John M. 2009. “The Effect of Gene Drive on Containment of Transgenic Mosquitoes.” Journal of Theoretical Biology 258 (2): 250–265. doi:10.1016/j.jtbi.2009.01.031.
- Marshall, John M., Anna Buchman, Héctor M. Sánchez C, and Omar S. Akbari. 2017. “Overcoming Evolved Resistance to Population-Suppressing Homing-Based Gene Drives.” Scientific Reports 7 (1): 921. doi:10.1038/s41598-017-02744-7 doi: 10.1038/s41598-017-01037-3
- Marshall, John M., and Bruce A. Hay. 2012. “Confinement of Gene Drive Systems to Local Populations: A Comparative Analysis.” Journal of Theoretical Biology 294 (February): 153–171. doi:10.1016/j.jtbi.2011.10.032.
- Matthews, Benjamin J., Carolyn S. McBride, Matthew DeGennaro, Orion Despo, and Leslie B. Vosshall. 2016. “The Neurotranscriptome of the Aedes Aegypti Mosquito.” BMC Genomics 17: 32. doi:10.1101/026823 doi: 10.1186/s12864-015-2239-0
- Min, John, Charleston Noble, Devora Najjar, and Kevin Esvelt. 2017a. “Daisy Quorum Drives for the Genetic Restoration of Wild Populations.” BioRxiv, 115618. doi:10.1101/115618.
- Min, John, Charleston Noble, Devora Najjar, and Kevin M. Esvelt. 2017b. “Daisyfield Gene Drive Systems Harness Repeated Genomic Elements as a Generational Clock to Limit Spread.” BioRxiv, 104877. doi:10.1101/104877.
- Muscarella, D. E., and V. M. Vogt. 1993. “A Mobile Group I Intron from Physarum Polycephalum Can Insert Itself and Induce Point Mutations in the Nuclear Ribosomal DNA of Saccharomyces Cerevisiae.” Molecular and Cellular Biology 13 (2): 1023–1033. doi: 10.1128/MCB.13.2.1023
- Noble, Charleston, John Min, Jason Olejarz, Joanna Buchthal, Alejandro Chavez, Andrea L. Smidler, Erika A. DeBenedictis, George M. Church, Martin A. Nowak, and Kevin M. Esvelt. 2016. “Daisy-Chain Gene Drives for the Alteration of Local Populations.” BioRxiv, 057307. doi:10.1101/057307.
- Noble, Charleston, Jason Olejarz, Kevin M. Esvelt, George M. Church, and Martin A. Nowak. 2017. “Evolutionary Dynamics of CRISPR Gene Drives.” Science Advances 3 (4): e1601964. doi:10.1126/sciadv.1601964.
- O’Brochta, David A., Nagaraja Sethuraman, Raymond Wilson, Robert H. Hice, Alexandra C. Pinkerton, Cynthia S. Levesque, Dennis K. Bideshi, et al. 2003. “Gene Vector and Transposable Element Behavior in Mosquitoes.” The Journal of Experimental Biology 206 (Pt 21): 3823–3834. doi: 10.1242/jeb.00638
- O’Hara, Peter. 2006. “The Illegal Introduction of Rabbit Haemorrhagic Disease Virus in New Zealand.” Revue Scientifique et Technique (International Office of Epizootics) 25 (1): 119–123.
- Oye, Kenneth A., Kevin Esvelt, Evan Appleton, Flaminia Catteruccia, George Church, Todd Kuiken, Shlomiya Bar-Yam Lightfoot, Julie McNamara, Andrea Smidler, and James P. Collins. 2014. “Regulating Gene Drives.” Science (New York, N.Y.) 345 (6197): 626–628. doi:10.1126/science.1254287.
- Paine, R. T. 1969. “A Note on Trophic Complexity and Community Stability.” The American Naturalist 103 (929): 91–93. doi:10.1086/282586.
- Petersen, Alexander M., Massimo Riccaboni, H. Eugene Stanley, and Fabio Pammolli. 2012. “Persistence and Uncertainty in the Academic Career.” Proceedings of the National Academy of Sciences 109 (14): 5213–5218. doi:10.1073/pnas.1121429109.
- Port, Fillip, Hui-Min Chen, Tzumin Lee, and Simon L. Bullock. 2014. “Optimized CRISPR/Cas Tools for Efficient Germline and Somatic Genome Engineering in Drosophila.” Proceedings of the National Academy of Sciences of the United States of America 111 (29): E2967–E2976. doi:10.1073/pnas.1405500111.
- Prowse, Thomas A. A., Phillip Cassey, Joshua V. Ross, Chandran Pfitzner, Talia A. Wittmann, and Paul Thomas. 2017. “Dodging Silver Bullets: Good CRISPR Gene-Drive Design Is Critical for Eradicating Exotic Vertebrates.” Proceedings. Biological Sciences 284 (1860), doi:10.1098/rspb.2017.0799.
- Rasgon, Jason L. 2009. “Multi-Locus Assortment (MLA) for Transgene Dispersal and Elimination in Mosquito Populations.” PLoS ONE 4 (6): e5833. doi:10.1371/journal.pone.0005833.
- Reeves, R. Guy, Jarosław Bryk, Philipp M. Altrock, Jai A. Denton, Floyd A. Reed, and Alexander W. E. Franz. 2014. “First Steps Towards Underdominant Genetic Transformation of Insect Populations.” PloS One 9 (5): e97557. doi:10.1371/journal.pone.0097557.
- Renn, Ortwin, and Debra Levine. 1991. “Credibility and Trust in Risk Communication.” In Communicating Risks to the Public, edited by Roger E. Kasperson and Pieter Jan M. Stallen, 175–217. Technology, Risk, and Society 4. Springer. doi:10.1007/978-94-009-1952-5_10.
- Roberts, Andrew, Paulo Paes de Andrade, Fredros Okumu, Hector Quemada, Moussa Savadogo, Jerome Amir Singh, and Stephanie James. 2017. “Results from the Workshop ‘Problem Formulation for the Use of Gene Drive in Mosquitoes.’” The American Journal of Tropical Medicine and Hygiene 96 (3): 530–533. doi: 10.4269/ajtmh.16-0726
- Serebrovskii, A. S. 1940. “On the Possibility of a New Method for the Control of Insect Pests.” Zoologicheskii Zhurnal 19: 618–680.
- Simoni, Alekos, Carla Siniscalchi, Yuk-Sang Chan, David S. Huen, Steven Russell, Nikolai Windbichler, and Andrea Crisanti. 2014. “Development of Synthetic Selfish Elements Based on Modular Nucleases in Drosophila Melanogaster.” Nucleic Acids Research, doi:10.1093/nar/gku387.
- Sinkins, Steven P., and Fred Gould. 2006. “Gene Drive Systems for Insect Disease Vectors.” Nature Reviews. Genetics 7 (6): 427–435. doi:10.1038/nrg1870.
- Spradling, Allan. 2015. “Gene Drive: More Research, Not More Regulations.” Genes to Genomes, November 24. https://genestogenomes.org/gene-drive-more-research-not-more-regulations/.
- Sugahara, Ryohei, Shinjiro Saeki, Akiya Jouraku, Takahiro Shiotsuki, and Seiji Tanaka. 2015. “Knockdown of the Corazonin Gene Reveals Its Critical Role in the Control of Gregarious Characteristics in the Desert Locust.” Journal of Insect Physiology 79 (August): 80–87. doi:10.1016/j.jinsphys.2015.06.009.
- Thyme, Summer B., Sandrine J. S. Boissel, S. Arshiya Quadri, Tony Nolan, Dean A. Baker, Rachel U. Park, Lara Kusak, Justin Ashworth, and David Baker. 2014. “Reprogramming Homing Endonuclease Specificity Through Computational Design and Directed Evolution.” Nucleic Acids Research 42 (4): 2564–2576. doi:10.1093/nar/gkt1212.
- Tijdink, J. K., K. Schipper, L. M. Bouter, P. Maclaine Pont, J. de Jonge, and Y. M. Smulders. 2016. “How Do Scientists Perceive the Current Publication Culture? A Qualitative Focus Group Interview Study among Dutch Biomedical Researchers.” BMJ Open 6 (2): e008681. doi:10.1136/bmjopen-2015-008681.
- Torres, R., M. C. Martin, A. Garcia, Juan C. Cigudosa, J. C. Ramirez, and S. Rodriguez-Perales. 2014. “Engineering Human Tumour-associated Chromosomal Translocations with the RNA-Guided CRISPR-Cas9 System.” Nature Communications 5: 233. doi:10.1038/ncomms4964.
- Tsao, Jean I., J. Timothy Wootton, Jonas Bunikis, Maria Gabriela Luna, Durland Fish, and Alan G. Barbour. 2004. “An Ecological Approach to Preventing Human Infection: Vaccinating Wild Mouse Reservoirs Intervenes in the Lyme Disease Cycle.” Proceedings of the National Academy of Sciences of the United States of America 101 (52): 18159–18164. doi:10.1073/pnas.0405763102.
- Vanderplank, F. L. 1944. “Hybridization Between Glossina Species and Suggested New Method for Control of Certain Species of Tsetse : Abstract : Nature.” Nature 154: 607–608. doi:10.1038/154607a0.
- Vanderplank, F. L. 1948. “Experiments in Crossbreeding Tsetse-Flies, Glossina Species.” Annals of Tropical Medicine and Parasitology 42 (2): 131–152. doi: 10.1080/00034983.1948.11685357
- Wade, M. J., and R. W. Beeman. 1994. “The Population Dynamics of Maternal-Effect Selfish Genes.” Genetics 138 (4): 1309–1314.
- WHO. 2014. “TDR | The Guidance Framework for Testing Genetically Modified Mosquitoes.” http://www.who.int/tdr/publications/year/2014/guide-fmrk-gm-mosquit/en/.
- Wilson, James M. 2009. “A History Lesson for Stem Cells.” Science 324 (5928): 727–728. doi:10.1126/science.1174935.
- Windbichler, Nikolai, Philippos Aris Papathanos, and Andrea Crisanti. 2008. “Targeting the X Chromosome During Spermatogenesis Induces Y Chromosome Transmission Ratio Distortion and Early Dominant Embryo Lethality in Anopheles Gambiae.” PLoS Genetics 4 (12): e1000291. doi:10.1371/journal.pgen.1000291.
- Wood, R. J., L. M. Cook, A. Hamilton, and A. Whitelaw. 1977. “Transporting the Marker Gene Re (Red Eye) into a Laboratory Cage Population of Aedes Aegypti (Diptera: Culicidae), Using Meiotic Drive at the MD Locus.” Journal of Medical Entomology 14 (4): 461–464. doi: 10.1093/jmedent/14.4.461
