?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Gastrointestinal tract is a series of hollow organs connected in a long tube twisting from the mouth to the anus. Recovery of gastrointestinal diseased patients depends on the early diagnosis of the disease and proper treatment. In recent years, the diagnosis of gastrointestinal tract diseases using endoscopic image classification has become an active area of research in the biomedical domain. However, previous studies show that there is a need for improvement as some classes are more difficult to identify than others. In this study, we propose a concatenated neural network model by concatenating the extracted features of VGGNet and InceptionNet networks to develop a gastrointestinal disease diagnosis model. The deep convolutional neural networks VGGNet and InceptionNet are trained and used to extract features from the given endoscopic images. These extracted features are then concatenated and classified using machine learning classification techniques (Softmax, k-Nearest Neighbor, Random Forest, and Support Vector Machine). Among these techniques, support vector machine (SVM) achieved the best performance compared to others, using the available standard dataset. The proposed model achieves a classification accuracy of 98% and Matthews’s Correlation Coefficient of 97.8%, which is a significant improvement over previous techniques and other neural network architectures.
PUBLIC INTEREST STATEMENT
The digestive system consists of the gastrointestinal tract and other organs which help the body to break down and absorb food. The gastrointestinal tract may be affected by a variety of diseases which affect its functionality. The domain of gastrointestinal endoscopy includes the endoscopic diagnosis of various digestive diseases using image analysis and various devices. Endoscopy is currently the preferred method for examining the gastrointestinal tract; however, previous studies have shown that there is a need for improvement as some classes are more difficult to identify than others. In this study, we proposed a concatenated neural network model by concatenating the extracted features of VGGNet and InceptionNet networks to develop a gastrointestinal disease diagnosis model. The deep convolutional neural networks VGGNet and InceptionNet are trained and used to extract features from the given endoscopy images. The proposed model achieves a classification accuracy of 98% and Matthews’s Correlation Coefficient of 97.8%, which is a significant improvement over previous techniques and other neural network architectures.
1. Introduction
The human gastrointestinal tract is prone to several abnormal outcomes from minor discomfort to life-threatening diseases (Naz et al., Citation2021). Gastrointestinal tract diseases such as ulcers, bleeding, polyps, Crohn’s disease, colorectal cancer, tumor cancer, and other related diseases are well known today worldwide (Jha et al., Citation2021). According to the International Agency for Research on Cancer, the number of new cases of gastrointestinal cancer and the number of related deaths worldwide in 2018 is estimated at 4.8 million and is responsible for 26% of global cancer cases and 35% of all cancer deaths (Naz et al., Citation2021). Early diagnosis of various gastrointestinal diseases can result in effective treatment and reduce the risk of mortality. Unfortunately, various gastrointestinal diseases are undetectable or create confusion during screening by medical experts due to noise in the images which suppress important details (Borgli et al., Citation2020; Khan, Majid et al., Citation2021; Ramzan et al., Citation2021). Visual assessment of endoscopy images is subjective, often time-consuming, and minimally repetitive, which could lead to an inaccurate diagnosis (Borgli et al., Citation2020). The use of artificial intelligence (AI) in a variety of gastrointestinal endoscopic applications has the potential to improve clinical practice and improve the efficiency and precision of current diagnostic methods (Chanie et al., Citation2020; Woreta et al., Citation2015). Several studies have been conducted to classify gastrointestinal diseases with the goal of making a significant contribution to the effective diagnosis and treatment of gastrointestinal diseases. However, several studies used a small amount of data , and the majority of research studies aimed to classify a small number of gastrointestinal disorders in a specific part of the human gastrointestinal system. However, in Borgli et al. (Citation2020), a series of experiments were conducted based on deep convolutional neural networks to classify 23 different classes of images. The authors achieved a Matthews Correlation Coefficient of 0.902. More importantly, the study showed that there is a need for improvement as some gastrointestinal diseases are more difficult to identify than others.
Deep learning techniques are another form of machine learning techniques which have been used in several areas of gastrointestinal endoscopy including colorectal polyp detection and classification, analysis of endoscopic images for diagnosis of helicobacter pylori infection detection and depth assessment of early gastric cancer, and detection of various abnormalities in wireless capsule endoscopy images (Khan, Nasir et al., Citation2021; Khan, Sarfraz et al., Citation2020; Majid et al., Citation2020). Öztürk & Özkaya (Citation2020) stated that due to the deficiencies of radiologists and other human factors which could lead to a false diagnosis, a computer-aided automated system would be useful for accurately diagnosing gastrointestinal polyps in the early stages of cancer. In this paper, we proposed a gastrointestinal disease diagnosis model that uses SVM as a classifier to determine which of the 23 gastrointestinal diseases the endoscopic scan images correspond to. The main contributions of this paper are as follows:
We propose a novel concatenated feature extraction model of the VGGNet and InceptionNet deep learning models for endoscopic image classification. This method builds on convolving images using concatenated VGGNET and InceptionNet architectures for feature modeling and then classifying these features using RBF kernel-based multi-support vector machine. The proposed approach avoids the use of handcrafted feature extraction and selection. Furthermore, the proposed model overcomes the problem of over-fitting and requires few parameters to learn discriminant features.
We consider two popular pre-trained CNN models with transfer learning principles for robustness testing of the proposed model.
The remainder of this paper is organized as follows. Section 2 presents an overview of related works. Section 3 explains the materials and methods, while Section 4 articulates the experimental results and comparative analysis. Finally, Section 5 deals with the concluding remarks.
2. Related works
Over the past few years, the strength of deep learning algorithms have been explored in the field of endoscopy including capsule endoscopy, esophagus-astro-duodenoscopy, and colonoscopy to help doctors successfully diagnose various gastrointestinal lesions (Borgli et al., Citation2020; Chanie et al., Citation2020; Khan, Nasir et al., Citation2021; Khan, Khan et al., Citation2020; Gamage et al., Citation2019). Nowadays, deep learning approaches especially CNN has become a powerful machine learning technique in image processing tasks such as the classification of gastrointestinal diseases.
Borgli et al. (Citation2020) carried out a series of experiments based on deep convolutional neural networks using applied common architectures with a slight modification to classify 23 different classes of images with the help of Pre-Trained ResNet-50, Pre-Trained ResNet-152, Pre-Trained DensNet-161, Averaged ResNet-152 + DenseNet-161, and ResNet-152 + DenseNet-161 + MLP which achieve a Matthews correlation coefficient (MCC) of 0.826, 0.898, 0.899, 0.899, and 0.902, respectively. However, the results also show that there is room for improvement as some classes are more difficult to identify than others (difficulty in classifying ulcerative colitis and esophagitis, difficulty in classifying dyed lifted polyps and dyed resection margins, and difficulty in classifyingbarrettes from esophagitis or z-line).
The descriptive study of Woreta et al. (Citation2015) conducted at Gondar University Hospital based on patient data indicates that esophago-gastroduodenal pathology was detected in 1093 (83.4%) patients of which duodenal ulcer was the most common endoscopic findings obtained from 333 (25.4%) cases.
The study performed by Chanie et al. (Citation2020) at Saint Paul’s Hospital Millennium Medical College on 128 patients varices indicates that the most common cause of upper gastrointestinal bleeding was observed in 46.1% (59), followed by peptic ulcer disease, 24.2% (31), esophagitis, 3.9% (5), gastritis, 6.3% (8), duodenitis, 3.1% (4), and malignancy, 4.7% (6), while 10 patients (7.8%) had both varices and ulcer. The proportion of deaths in the study was 17.2%.
Khan, Nasir et al., (Citation2021) presented a ulcer segmentation approach based on deep learning of the modified Recurrent CNN masks for the detection of ulcers and classification of gastrointestinal diseases (ulcer, polyp, bleeding). During the classification phase, the ResNet101 pre-trained CNN model is further developed through transfer learning to derive deep characteristics. Grasshopper optimization and the minimal distance fitness function were used to improve the acquired deep features. For the final classification, the best-selected features are fed into a multi-support vector machine classifier using the cubic kernel function. Validation is done with a private data set, which achieves a MOC of 88.08% for ulcer segmentation and 99.92% for the classification accuracy. The ulcer regions were not correctly segmented because of the small amount of training data which lead to the failure of the approach to segment polyp and bleeding regions.
Gamage et al. (Citation2019) used pre-trained DenseNet-201, ResNet-18, and VGG-16 CNN models as feature extractors with a global average pooling (GAP) layer to produce an ensemble of deep features as a single feature vector with a promising accuracy of 97.38%. Only eight classes of digestive tract anomalies were predicted by the suggested method.
Takiyama et al. (Citation2018) proposed an alternative CNN-based classification model for categorizing the anatomical location of the human digestive tract. This technique could classify EGD images into four large anatomical locations (larynx, esophagus, stomach, and duodenum) and three subcategories for stomach images (upper, middle, and lower regions). A predetermined CNN architecture called GoogleNet was used for this classification problem, which achieved a high performance.
Owais et al. (Citation2019) proposed a CNN and TML classification framework for the classification of several gastrointestinal diseases using endoscopic videos with a dataset containing a total of 77 video files with52,471 images. They considered a total of 37 different classes (both diseased and normal cases) related to the human GI tract and achieved an average accuracy of 92.57%. However, more data was required to improve the systems performance and due to the limited size of the dataset, it was not possible to use different videos for training and testing, because most of the classes (about 21 out of 37 classes) consisted of single-patient data (one video per class).
Sharif et al. (Citation2019) proposed a new technique based on merging deep CNN and geometrical functions. Initially, the disease regions are extracted from the given WCE images using a new approach called improved contrast color features. The geometric features were derived from the segmented disease area. Thereafter, a unique fusion of the deep CNN functions, VGG16 and VGG19 was carried out based on the Euclidean Fisher Vector. The unique features are combined with geometric features which are then used to choose the best features using the conditional entropy approach. K-Nearest Neighbor (kNN) was used to classify the selected features. For the evaluation of the suggested method, a privately gathered database of 5500 WCE images was used which achieved a classification accuracy of 99.42% and a precision rate of 99.51%. However, the authors obtained just three classification classes, namely: ulcers, bleeding, and health.
Shichijo et al. (Citation2017) proposed a deep learning CAD tool for the diagnosis of Helicobacter pylori (H. pylori) infection. The proposed framework employed two-step CNN models. In the first step, a 22-layer deep CNN was refined for classification (i.e., positive or negative) of H. pylori infection. Then, in step 2, another CNN was used to further classify the data set (EGD images) according to eight different anatomical locations to achieve an accuracy of 83.1%.
Segu et al. (Citation2016) introduced a deep CNN system for characterizing small intestine motility. This CNN-based method exploited the general representation of six different intestinal motility events by extracting deep features, which resulted in superior classification performance when compared to the other handcrafted feature-based methods. Although they achieved a high classification performance of 96% accuracy, they only considered a limited number of classes.
Li et al. (Citation2012) used machine learning methods to analyze lymph node metastasis in gastric cancer using gemstone spectral imaging. They used feature selection and metric learning methods to reduce data dimension and feature space, and then used the kNN classifier to distinguish lymph node metastasis from non-lymph node metastasis using 38 lymph node samples in gastric cancer to achieve an overall accuracy of 96.33%. Wang et al. (Citation2015) proposed a system for detecting polyps in colonoscopy. It can generate an alert with real-time feedback during colonoscopy. The authors used visual elements and a rule-based classifier to detect polyp borders. The system achieved an accuracy of 97.7% for polyp detection.
A multiscale approach for detecting ulcers was introduced by Souaidi et al. (Citation2019). The authors retrieved texture patterns such as the entire pyramids of LBP and Laplacian, and then classified them using SVM. They tested the system on two WCE datasets and found that it achieved accuracies of 95.11% and 93.88%. Li & Meng (Citation2012) proposed a framework for wireless CE images through integrated LBP and discrete wave transformation to extract texture characteristics from scale and rotation invariants. Finally, the selected characteristics were categorized using the SVM classifier.
By combining the bag-of-features (BoF) approach with the salience map, Yuan & Meng (Citation2014) proposed an integrated polyp identification system. The bag-of-features approach uses a Scale-Invariant Feature Transformation feature vector with k-means clustering to characterize local features in the first stage. The saliency map was then used to generate a histogram, which was then used to extract saliency features. Finally, the BoF and primary attributes were sent into the SVM classifier, which was used to classify the data. For the detection of polyps and ulcers in the small intestine in CE, Karargyris & Bourbakis (Citation2011) proposed a geometric and texture-based technique. The images were pre-processed with Log Gabor filters and the SUSAN edge detector, and geometrical features were retrieved to determine the polyp and ulcer area. To identify normal and pathological tissues, Li & Meng (Citation2012) employed discrete wavelet processing and a uniform local binary model (LBP) with SVM. Wavelet transform combines the ability of multi-resolution and uniform LBP analysis in this feature extraction approach to give robustness to lighting fluctuations, resulting in superior performance.
Most of the presented approaches have been designed to classify a limited number of gastrointestinal diseases in a specific portion of the human gastrointestinal tract. Moreover, feature extraction is a challenging task in medical imaging for recognition of infections because of additional and irrelevant features, or noises that degrades the overall recognition accuracy. Also, some gastrointestinal diseases are difficult to identify from the other disease types. Therefore, to deal with these challenges, in this paper, we employ different image processing stages. Finally, we used deep feature concatenation as a single feature vector by combining VGGNet and InceptionNet models as feature extractors, followed by a SVM classification.
3. Material and methods
3.1. Dataset acquisition
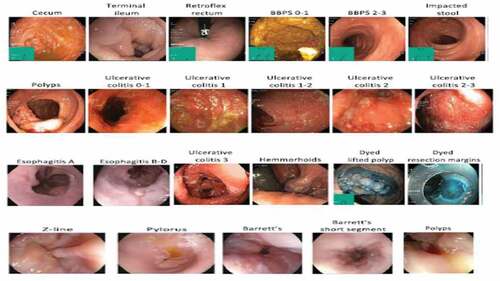
In this paper, we used the publicly available HyperKvasir Dataset (Borgli et al., Citation2020). The dataset is composed of endoscopic imagery from inside the GIT which has 23 different classes. It contains anatomical landmarks such as cecum, ileum, retroflex-rectum, pylorus, retroflex-stomach, z-line, and pathological findings such as hemorrhoids, polyps, ulcerative-colitis-grade-0-1, ulcerative colitis grade-1, ulcerative colitis grade-1-2, ulcerative colitis grade-2, ulcerative colitis grade-2-3, ulcerative colitis grade-3, Barrett’s, short segment Barrett, esophagitis-a, esophagitis b-d, and quality of mucosal views which include bbps 0–1, bbps 2–3, and impacted stool and therapeutic interventions; dyed-lifted-polyps, dyed-resection-margins. In total, the dataset contains 10,662 labeled images stored using the JPEG format. Finally, the data is split into 70/30 which means 70% for training, 20% for testing, and 10% of the data for validation. Image samples of the various labeled classes for gastrointestinal disease are shown in .
3.2. Proposed methodology
The proposed gastrointestinal disease classification model development comprises of different processes such as preprocessing, segmentation, feature extraction, and classification.
3.2.1. Preprocessing
Each endoscopy image is stored, and its quality might be degraded by different factors such as no uniform intensity, variations, motions, shift, and noise. Thus, it requires preprocessing techniques to reduce the disease identification complexity. In this study, we used image preprocessing techniques such as image resizing, and noise filtering techniques.
3.2.1.1. Image resizing
The dataset has images that have a uniform size of 100 × 100 resolution, but due to our computer performance, the image resolution of the original image needed to be resized before feeding it into the model. Therefore, the size of the image was resized to a resolution of 32 × 32.
3.2.1.2. Denoising
Medical images play a vital role in disseminating information about polyps, cancers, hemorrhoids, and other diseases. A major challenge in the process of medical imaging is to obtain an image without the loss of any significant information. The images obtained were affected by noise, and this noise affects the classification accuracy of the proposed model. Basic digital image filters like Gaussian filter (GF), Median filter (MF), and Adaptive Median filter (AMF) (Guan et al., Citation2014) are common filters used to remove noise in medical images. In this study, we compared GF, MF, and AMF todetermine the most suitable filtering technique that improves the classification performance of the proposed model.
i. Adaptive median filter
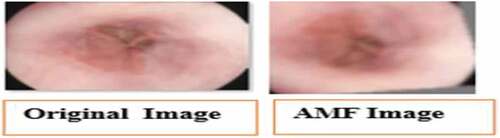
Adaptive median filter (AMF) is being applied widely as an advanced denoising technique compared with the traditional median filter because the AMF executes spatial processing to determine which pixels in the image has been affected by noise. It classifies pixels by comparison of each pixel in the image to its surrounding neighbor pixel. It preserves image details such as edges and smoothens non-impulsive noise, while the standard median filter does not (Ali, Citation2018). An illustration of the resultant image after using the noise removal technique is shown in .
3.2.1.3. Data augmentation
Deep learning has been used to classify several types of medical images over the past few years and has achieved a desirable performance. This was found to be true when there is an extensive amount of data that helps to attain the desired performance model. The limitations of the lack of training data are addressed through augmentation and transfer learning, which is very essential because of the inadequate medical datasets. Data augmentation enhances classification performance and prevents the overfitting issue since it offers a large amount of data (Shorten & Khoshgoftaar, Citation2019). In this paper, we applied rotation of 45°, width_shift_range = 0.2, height_shift_range = 0.2, and horizontal flip augmentation technique. After augmentation, the dataset is increased to 47,398 images.
3.2.2. Feature extraction
Feature extraction is a type of dimensionality reduction where a large number of pixels of an image are efficiently represented in such a way that essential parts of the image are captured effectively. For feature extraction, a concatenated model of VGGNet and the InceptionNet CNN architectures was used. The concatenation process of the two model features integrates the information from the different CNNs to create a more discriminative feature representation than when only the feature extracted from a single CNN model is used since different CNN architectures can capture diverse information of input images which increase their performance compared to a single model (Lonseko et al., Citation2021). The concatenation of both model features is taken as the input to the next classifier.
3.2.2.1. Feature extraction using the proposed VGGNet model
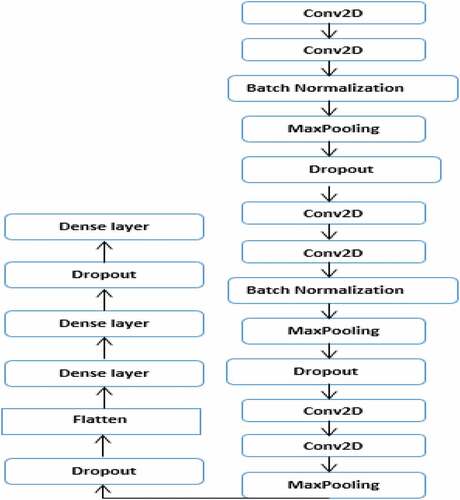
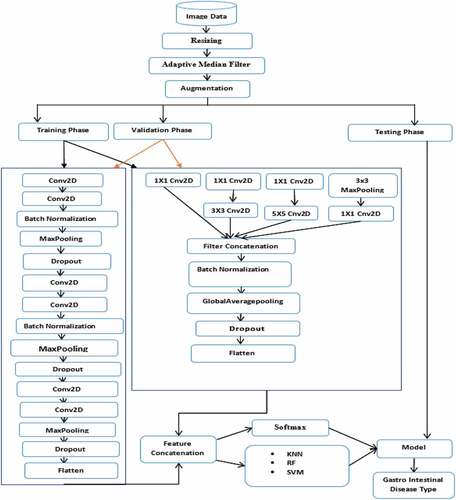
In this study, high-level features are extracted using the proposed VGGNet model. Despite this, feature extraction with CNN models consists of several similar steps because each step is made up of cascading layers: convolution, pooling, dropout, batch normalization, and dense layer, each of which represents features at different levels of abstraction. However, CNN models are different from each other in how these important layers are adjusted and also in how they form the network. The architectural design of the proposed VGGNet model used in this study is presented in , while the parameters used for the proposed VGGNet architecture of the CNN model are presented in . The proposed VGGNet model comprises of six convolutions, two batch normalizations, three max-pooling, three dropouts, and one flattened layer. A dropout layer is inserted below each max pooling and dense layer to overcome overfitting. Finally, the extracted features are classified into 23 gastrointestinal disease types by Softmax, KNN, RF, and SVM classifiers. When we trained the model, we observed overfitting of the training data. During this time, the model performs well with the training set but didnt perform well with the validation or test set. To solve this challenge, we used data augmentation to increase the dataset and dropout operation for regularization.
Table 1. Parameters of the proposed VGGNet architecture of the CNN model
3.2.2.2. Feature extraction using the proposed InceptionNet model
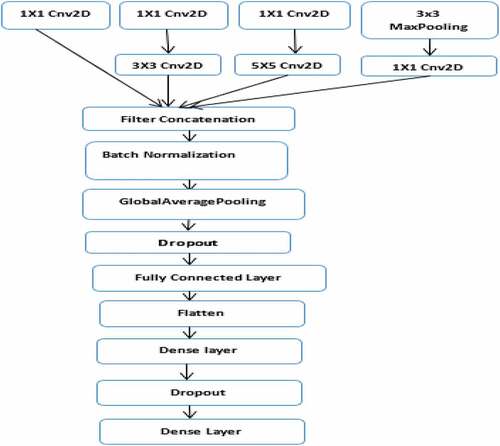
The second model (the architectural design of the proposed InceptionNet model) used in this study is described in , while the parameters used for the proposed InceptionNet architecture of the CNN model are presented in . The model comprises of six convolutions, one max-pooling, one global average polling, one batch normalization, one dropout, and one flattened layer. A dropout layer is inserted below the global average pooling and dense layer to overcome overfitting. Finally, the extracted features are classified into 23 gastrointestinal disease types using Softmax, KNN, RF, and SVM classifiers. When we trained the model, we observed overfitting of the training data in the same way as the other model. This was solved by adding a dropout layer.
Table 2. Description of the parameters of the proposed InceptionNet architecture of the CNN model
3.2.2.3. Feature extraction using the proposed concatenated model
As previously stated, the proposed model aims to accurately diagnose gastrointestinal disease by concatenating deep features extracted from endoscopy images by using two different models (CNN architecture of VGGNet and InceptionNet). First, a VGGNet model is proposed to extract features from endoscopy images. The proposed InceptionNet model extracts features from the same images in the other model. Finally, the extracted features from these models are flattened and concatenated into a single classification descriptor. Finally, the extracted features are fed into the classifier.
3.2.3. Radial Basis Function (RBF) kernel-based SVM classifier
Classification is the final step of the proposed gastrointestinal disease classification model. The features which are extracted and concatenated from the two models are classified as either cecum, ileum, retroflex-rectum, pylorus, retroflex-stomach, z-line, bbps 0–1, bbps 2–3, impacted-stool, hemorrhoids, polyps, ulcerative colitis grade-0-1, ulcerative colitis grade-1, ulcerative colitis grade-1–2, ulcerative colitis grade-2, ulcerative colitis grade-2–3, ulcerative colitis grade-3, barrett’s, short segment Barrett, esophagitis a, esophagitis b-d, dyed-lifted-polyps, or dyed resection margins. In this study, we compared four classifiers, Softmax, KNN, RF, and SVM to determine the best classifier for correctly identifying gastrointestinal diseases using the proposed model. A Radial Basis Function kernel-based SVM classifier with a concatenated feature extraction model was also proposed to efficiently classify the type of gastrointestinal disease. Support Vector Machine is a supervised classification algorithm presented in 1992 by Boser, Guyon, and Vapnik and it has been broadly utilized for classification, regression, detection tasks, and feature reduction (Paoletti et al., Citation2020). Support vector machine (SVM) tries to find the optimal hyperplane which can be used to classify new data points into two classes with an ideal isolating hyperplane by increasing the distance between the nearest vectors to the hyperplane (Aychew, Citation2020). After breaking down the multiclass classification problem into multiple binary classification problems, the same principle is used for multiclass classification as well as binary classification. Then, the algorithm can classify an instance as only one of three classes or more. Different SVM algorithms use different types of kernel functions like—Polynomial Kernel, Gaussian Kernel, Radial Basis Function (RBF), Sigmoid Kernel, and others. The most used type of kernel function is RBF because it has localized and finite responses along the entire x-axis (Salau & Jain, Citation2021). Because real noisy data cannot be linearly separated in this case, SVM is preferable because it can still be used for nonlinear classification problems by applying appropriate kernel functions that map the input data into a higher-dimensional space where the classes are linearly separable (Jain & Salau, Citation2019), and when the amount of available training data is small, the SVM classifier outperforms the softmax classifier for the CNN features (Ayalew et al., Citation2022). RBF is used in this study to solve our 23 class classification problem because it is the most commonly used SVM kernel function used by several researchers. We used C values of 1, 10, 100, and 1000, as well as gamma values of 0.001 and 0.0001. The best model performance was obtained when the c value was 1 and the gamma value was 0.001.
3.2.4. Convolutional neural network
Computer vision can be very useful in practical areas such as surgery, where it can improve surgeon performance by providing real-time contextual awareness, and endoscopy, where it can provide proper therapy after surgery (Maier-hein et al., Citation2017). Machine perception covers a range of levels, from low-level tasks like identifying edges, to high-level tasks like understanding full scenes because of the maturation of deep learning and the supply of large datasets labeled to train these algorithms (Salau & Jain, Citation2019; Deng et al., Citation2009). When there is a lack of domain understanding for feature extraction, CNN is preferable because useful learned features are extracted automatically rather than handcrafted based algorithms that manually define and extract a set of features such as edge detection, corner detection, histograms, and so on. However, this approach does not guarantee that the number of corners is a good descriptor for classifying images (Noor Mohamed & Srinivasan, Citation2020; Lin et al., Citation2020). Deep Learning is now widely used to solve complex problems like image classification, natural language processing, and voice recognition. RBM, Autoencoder, DBN, CNN, and RNN are the most important deep learning algorithms (Shetty & Siddiqa, Citation2019). According to Guo et al. (Citation2016), CNN-based architectures are most widely used for image classification problems. CNN is currently the most widely used machine learning algorithm in medical image analysis because it preserves the spatial relationships when filtering input images, which is crucial in endoscopy, for example, where a normal colon tissue meets a cancerous tissue (Khan et al., Citation2020). CNN employs three basic types of neural layers: convolutional layers, pooling layers, and fully connected layers. Convolutional Layers: In the convolutional layers, CNN uses different portions of the image as computational intelligence to convolve the full image and produce different feature maps. Pooling layers: Pooling layers are in charge of reducing the spatial measurements (width, height) of the information volume for the next convolutional layer. This layer’s activity is also known as subsampling or downsampling, because the decrease in size results in a loss of data. Nonetheless, such loss is beneficial to the network because the reduction in size results in less computational overhead for the network’s subsequent layers, as well as the elimination of overfitting. The most commonly used techniques are average pooling and maximum pooling. Furthermore, there are various varieties of the pooling layers in literature, each propelled by different inspirations and serving specific needs, for example, stochastic pooling, spatial pyramid pooling, and def-pooling. Fully Connected Layers: After a few convolutional and pooling layers, the neural network’s important level of thinking is carried out through completely associated layers. As the name implies, neurons in a completely associated layer have complete associations with all representations in the previous layer. Their actuation can then be achieved by duplicating the framework followed by an inclination counterbalance. Finally, fully associated layers convert the two-dimensional component maps to a one-dimensional element vector. The obtained vector could either be carried forward into a specific number of classes for classification or considered as a component vector for additional preparation (Escobar et al., Citation2021). Pre-trained models are a type of deep learning models that have been trained on a large dataset to solve classification problems (Al-Adhaileh et al., Citation2021). VGGNet, ResNet, Inception, and Xception are some of the most recent basic deep learning structures for CNN. Simonyan and Zisserman introduced VGGNet in 2014. VGGNet was the first runner-up design in the characterization task of the ImageNet Large Scale Visual Recognition Competition (Simonyan & Zisserman, Citation2015; Sakib et al., Citation2018). ResNet CNN’s architecture demonstrates exceptional fundamental use of skip connections and batch normalization. Also, at the network’s end, there are no fully connected layers. The main flaw of this system is that due to the vastness of the boundaries, it is expensive to use. In any case, ResNet is currently regarded as the best convolutional neural network model, having won the ILSVRC 2015 challenge. It is worth noting that, despite having many more layers than the VGG, it requires much less memory, nearly 5 times less. This is because, instead of dense layers in the classification phase, this network employs GlobalAveragepooling, a type of layer that converts the final layer in the feature extraction stage’s two-dimensional activity maps to an n-class vector that is used to calculate the probability of belonging to each class. Szegedy et al. (Citation2015) proposed Inception, a deep convolutional neural network design that is responsible for setting a new critical edge for classification and detection in the ImageNet Large-Scale Visual Recognition Challenge in 2014 (ILSVRC14). The main advantage of this strategy is that it adds a critical component to a minor increase in computer requirements when compared to smaller and smaller networks. Chollet (Citation2017) designed Xception, which has 36 convolutional layers that serve as the foundation for extracting network features. The Xception architecture is made up of a linear stack of depth-separable convolution layers with residual connections. It is an adaptation of Inception, with deep-separable convolutions replacing the Inception modules. Furthermore, it has approximately the same number of parameters as Inception-v1. We used InceptionNet and VGGNet because both performed well in the 2014 ImageNet challenge, and InceptionNet gained attention due to its architecture based on modules called Inception.
3.3. Proposed CNN model
The proposed model aims to accurately diagnose gastrointestinal disease using concatenated deep features extracted from endoscopy images of two different models (CNN architecture of VGGNet and InceptionNet). Initially, the input images are appropriately prepared by resizing and noise filtering processes. The features extracted from endoscopy images are then extracted using the proposed VGGNet model. The proposed InceptionNet model extracts features from the same images in the other model. Finally, the features extracted from these models are combined into a single classification descriptor. depicts the proposed concatenated model architecture, which concatenates the features extracted by the two flattened models (CNN architecture of VGGNet and InceptionNet).
3.3.1. Architecture of the proposed model
The overall steps followed to identify the corresponding class of a particular input image are presented in the proposed gastrointestinal disease classification model architecture as shown in . There are three main phases in the systems architecture. The training phase, the testing phase, and the validation phase. The training phase and validation phase are performed before the testing phase for training the model. The testing phase is performed after the training phase and validation phase for testing the performance of the trained model with the new image dataset. First, the whole gastrointestinal image dataset is resized to 32 × 32. Secondly, the resized dataset is filtered with the AMF technique. Thirdly, filtered images are augmented. Then, the augmented images are subdivided into training dataset, validation dataset, and test dataset. Next, both the training dataset and validation dataset were fed to the concatenated model. The next step is feature extraction using the concatenated model of the VGGNet and InceptionNet architecture of the CNN model, which is used to extract the useful high-level features from the images. Lastly, extracted features of VGGNet and InceptionNet are concatenated together and fed into the classifiers (Softmax, KNN, RF, and SVM). While in the test set, both models features (VGGNet and InceptionNet) are concatenated together and fed to the previously trained SVM model. The detail of each phase of the proposed model is presented in section 3.3.2.
3.3.2. Transfer learning
Transfer learning is the process of improving learning in a new task by transferring knowledge from a previously learned related task. It is implemented by slicing off the final output layers of a pre-trained model and replacing them with output layers specific to the new target output. Pre-trained models, on the other hand, have been trained on an ImageNet dataset that contains 1000 classes of images, such as animals and cars. Because models trained on the ImageNet dataset learn features that are completely different from medical image features, fine-tuning the models for medical tasks may not have a significant impact on their performance.
Learning method
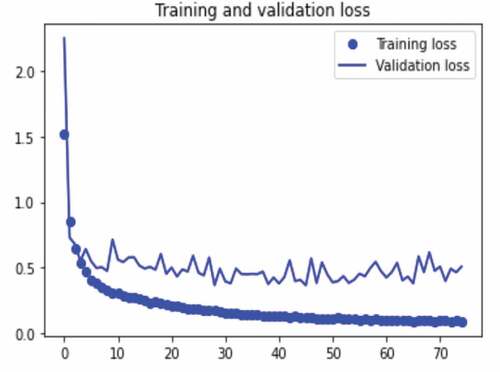
The proposed concatenated model is evaluated in terms of accuracy and loss function. To measure the difference between the output predictions and the target output, we used a categorical cross-entropy loss function, which is commonly used for multi-class problems. Adaptive momentum estimation (Adam) optimization function with an initial learning rate of 0.001 was used to adjust the learning rate and other parameters to increase the accuracy of the model and to decrease the loss. The classified classes are compared in accordance with the classification’s performance parameters. The confusion matrix is used to calculate the parameters performance. According to Ayalew et al. (Citation2022; Salau (Citation2021), each performance measure parameter is computed as follows:
False-Positive Rate (FPR): indicates that the diseases are not detected correctly.
False-Negative Rate (FNR): indicates that the diseases are detected as non-disease regions.
True Positive Rate (TPR): indicates that the diseases are detected as disease regions.
True Negative Rate (TNR): indicates that a non-disease region is correctly recognized as a non-disease region.
Confusion matrix (CM): CM is used to find the correctness and accuracy of the model. It is obtained using parameters such as TP, TN, FP, and FN of the matrix.
Accuracy: Accuracy is the number of correct predictions divided by the total number of samples. The formula for accuracy is given as follows:
Precision: It is a measure that indicates the proportion of patients that were diagnosed as having a disease. The predicted positives (People predicted with a disease are TP and FP) and the people having a disease are TP as follows:
Recall (Sensitivity): It is a measure that indicates the proportion of patients that have a disease and were diagnosed by the model as having a disease. The actual positives (People having a disease are TP and FN) and the people diagnosed by the model having a disease are TP as follows:
Dice similarity coefficient (F1-score): It represents a harmonic average of the precision and sensitivity and measures the similarity rate between predicted and ground-truth regions. It is calulated using Eq. (4):
Matthews Correlation Coefficient (MCC): accuracy is sensitive to class imbalance. Precision, recall, and F1-score are asymmetric, but MCC is perfectly symmetric; therefore, no class is more important than the other. It measures the classification quality even if the classes are of different sizes, true positives, and false positives and negatives as follows:
4. Experimental results and discussion
We compared different filtering and segmentation techniques, as well as different state-of-the-art classifiers and CNN pre-trained models with the proposed concatenated CNN model.
4.1. Experimental setup
Experiments were performed with the Keras prototype on the tensor flow backend. The performance of the proposed concatenated model is measured using accuracy, confusion matrix, precision, recall, f1-score, and Matthews correlation coefficient metrics of the classification. The whole image dataset before augmentation (10,662 images) or after augmentation (47,398 images) is partitioned into training, validation, and testing. Image augmentation is required to enable the trained model to extract a large number of important features. After augmentation, we used a dataset of 70% (33,944 images) for training, 20% (9610 images) for testing, and 10% (3844 images) for validation, with no overlap from the original images. The same splitting method was also applied to the dataset for the experiments which were performed before augmentation. For this study, we used an anaconda environment, python programming language, a free-source Keras and a free-source CNN library using Tensor Flow which served as the backend. The python code was written using Jupiter Notebook. The designed prototype was tested on the Windows 10 Hp Pavilion laptop with 64-bit operating system. The laptop is an Intel(R) Core (TM) i3-8145UMQ with 2.30 GHz CPU processor, with 8 GB RAM and 700 GB hard disk capacity.
4.2. Experiments before augmentation
In this stage, we trained the proposed concatenated model for 75 epochs, a batch size of 32, and an initial learning rate of 0.001. With the end-to-end proposed concatenated CNN model using a softmax classifier, we performed seven experiments; the first experiment was performed to obtain the result before noise filtering and segmentation techniques; the second and third experiment was performed to compare the three filtering techniques (gaussian filtering, median filtering, and adaptive median filtering). We have achieved 70.8%, 68.6%, and 72.7% accuracy, when we used GF, MF, and AMF, respectively. Since AMF achieves superior results, we employed AMF for the rest of the experiments and the last three experiments were performed for viewing the effect of the three segmentation techniques (we compared the otsu, adaptive thresholding, and kmeans segmentation techniques). We achieved 54.7%, 52.3%, and 56.3% accuracy when we used otsu, adaptive thresholding, and k-means, respectively. Since both segmentation techniques achieved low accuracies, we did not use segmentation techniques for the rest of the experiment. The results are shown in .
Table 3. Accuracy of the proposed model with resized data using noise filtering and segmentation techniques
4.3. Experiments after augmentation
In this stage, we conducted 14 experiments. We trained the proposed concatenated model using 75 epochs, a batch size of 32, and an initial learning rate of 0.001. The first four experiments were performed to compare the classifiers using the VGGNet feature extraction method. The results show that 93%, 93.2%, 93.3%, and 93.4% accuracy was achieved when we used Softmax, KNN, RF, and SVM, respectively. The second four experiments were performed to compare classifiers using the InceptionNet feature extraction method. The results show that 84%, 84.5%, 84.7%, and 85% accuracy was achieved when we used Softmax, KNN, RF, and SVM, respectively. The third and fourth experiments were performed to compare classifiers using concatenated VGGNet-InceptionNet feature extraction method. We achieved 94%, 95.4%, 95.7%, and 98% accuracy when we used Softmax, KNN, RF, and SVM, respectively. The last two experiments were used to demonstrate the performance of the pre-trained VGGNet16 and pre-trained Inceptionv3. The models achieve an accuracy of 89% and 91%, respectively. The results are shown in .
Table 4. Comparison of classifiers based on the proposed feature extraction methods
4.4. Performance of the proposed concatenated model using the SVM classifier
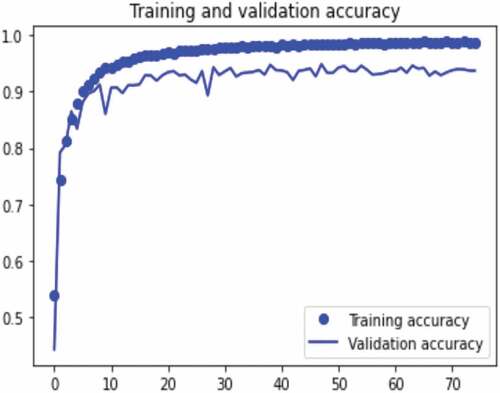
The results of the experiments performed to measure the training accuracy, validation accuracy, testing accuracy, and Matthews’s correlation coefficient of the proposed concatenated CNN model with SVM classifier are presented in . The experiments were performed using the concatenated model with a resized and a filtered (AMF technique) image dataset and SVM classifier. The experiments were performed using 75 epochs with a batch size of 32. We train the model with 33,944 training images and 3844 validation images, and we test with 9610 images. The model validation is done at the end of each epoch, while the model trains per epoch. At the end of the last epoch (epoch 75), our proposed concatenated model with SVM classifier obtains a training accuracy of 98.7%, validation accuracy of 98.2%, testing accuracy of 98%, and Matthews correlation coefficient of 97.8%. These results are shown in Figures .
Table 5. The precision, recall, training accuracy, validation accuracy, training loss, validation loss, Matthews correlation coefficient, and f1-score of the proposed concatenated CNN model with SVM classifier
4.5. Result discussion
The dataset was split into training, validation, and testing dataset for the proposed concatenated model training and the performance of the proposed concatenated model was tested using the dataset samples. To classify the gastrointestinal images into 23 classes, we have concatenated VGGNet and InceptionNet features for feature extraction and SVM for classification. During our experiment, we confirmed that the prediction ability of the model is increased when we used the concatenated features of the proposed VGGNet and InceptionNet models. Before using noise filtering and augmentation techniques, the accuracy of the model is 71.8%, and after noise removal, the accuracy of the model increased by 0.9% (72.7%). We obtained the best classification accuracy with the adaptive median noise filtering technique. We also applied the augmentation technique to control overfitting and to learn different features of images for the proposed concatenated model that helps to increase its classification ability. After augmentation, the accuracy of the proposed end-to-end model is 94%. We also applied different segmentation techniques to identify regions of interest in the image; however, these segmentation techniques result in the low classification performance of the model. Also, when the accuracy of each model was examined with the SVM classifier after the features were extracted with each of the feature extractors individually, the results show that VGGNet and InceptionNet have accuracies of 85% and 93.4%, respectively. Afterwards, the features of VGGNet and InceptionNet were concatenated which results in an improved classification performance of 98% accuracy. In this study, satisfactory results were obtained after we conducted 21 experiments. Finally, a comparison of state-of-the-art classifiers was performed using the proposed concatenated model. Our proposed concatenated model with SVM classifier achieves a training accuracy of 98.7%, validation accuracy of 98.2%, testing accuracy of 98%, and Matthews’s correlation coefficient of 97.8%. The results of the proposed concatenated model outperformed the proposed VGGNet, the proposed InceptionNet, and the state-of-the-art pre-trained VGGNet16 and Inceptionv3 models. Generally, based on the experiments, our proposed model improves Matthews’s correlation coefficient of the previous work by 7.6%. This indicates that the proposed concatenated model is effective for gastrointestinal disease diagnosis from endoscopy images. A comparative analysis of our proposed method with existing works is presented in . The results show that the proposed method achieves a high accuracy as compared to other methods.
Table 6. Comparison of proposed study with existing researches related to gastrointestinal disease
5. Conclusion
The digestive system consists of the gastrointestinal tract and other organs which help the body to break down and absorb food. The gastrointestinal tract is affected by a variety of diseases which makes it not to function properly. The domain of gastrointestinal endoscopy includes the endoscopic diagnosis of various digestive diseases using image analysis and various devices. Endoscopy is currently the preferred method for examining the gastrointestinal tract; however, its effectiveness is severely limited by the variation of the models performance. This study presents a concatenated model by concatenating features of VGGNet and InceptionNet for the classification of multiple gastrointestinal diseases from endoscopy images. The proposed model was trained and tested using a publicly available dataset from the HyperKvasir database. Extracted features were used to train the Softmax, KNN, RF, and SVM classifiers. The model achieves a test accuracy of 98% using the SVM classifier, which is an incredible performance when compared to state-of-the-art approaches. It outperforms the previous similar works by 7.6%, with Matthews’ correlation coefficient of 90.2%. As a result, the proposed model is one of the best gastrointestinal disease classification models. Based on the experimental outcomes of this study, we suggest the following recommendation as future work:
Segmentation plays a critical role in the early and accurate diagnosis of various diseases using computerized systems. For the future, we recommend that a new simultaneous segmentation method can be used by taking advantage of MASK-RCNN for detection of the disease location and GRABCUT for segmentation to improve the performance of the model.
This study was performed for only 23 gastrointestinal disease types; therefore, other types of gastrointestinal disease can also be considered.
In future works, it is important to evaluate different feature selection algorithms that can help to determine the smallest subset of features that can aid in the accurate classification of gastrointestinal disease types.
Consent for publication
The authors declare that they are in agreement with this submission and for the paper to be published if accepted.
Availability of data and material
All data generated or analyzed during this study are included in this published article.
Compliance with Journal Polices
The authors declare that they have no issues with journal polices and all journal policies have been abided by.
Details of any previous or concurrent submissions
This paper has not been submitted anywhere else for review.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
Notes on contributors
Ayodeji Olalekan Salau
Ayodeji Olalekan Salau received the M.Sc. and Ph.D. degrees in Electronic and Electrical Engineering from the Obafemi Awolowo University, Ile-Ife, Nigeria. His research interests include computer vision, artificial Intelligenceintelligence, and machine learning. Dr. Salau serves as a reviewer and editor for numerous reputable international journals. His research has been published in a number of reputable international conferences, book series, and major international journals. He is a recipient of the Quarterly Franklin Membership given by the Editorial Board of London Journals Press in 2020 for top qualitytop-quality research output. In 2020, Dr. Salau’s paper was awarded the best paper of the year 2019 in Cogent Engineering. In addition, he is the recipient of the International Research Award on New Science Inventions under the category of “Best Researcher Award” given by ScienceFather in 2020. In 2021, he was given the International Scientist Award on Engineering, Science, and Medicine. Presently, Dr. Salau works at Afe Babalola University in the Department of Electrical/Electronics and Computer Engineering.
References
- Al-Adhaileh, M. H., Ebrahim M. S., Fawaz W. A., Theyazn H. H Aldhyani, Nizar A., Ahmed A. A., Irfan U. M., Mohammed Y. A., Elham D. A., & Mukti E. J. (2021). Deep learning algorithms for detection and classification of gastrointestinal diseases. Complex, 2021, 6170416. https://doi.org/10.1155/2021/6170416.
- Ali, H. M. (2018). MRI Medical Image Denoising by fundamental filters. High-Resolution Neuroimaging - Basic Physical Principles and Clinical Applications. IntechOpen. https://doi.org/10.5772/intechopen.72427
- Ayalew, A. M., Salau, A. O., Abeje, B. T., & Enyew, B. (2022). Detection and classification of COVID-19 Disease from X-ray images using convolutional neural networks and histogram of oriented gradients. Biomedical Signal Processing and Control, 103530(74), 1–23. https://doi.org/10.1016/j.bspc.2022.103530
- Aychew, S. (2020). Designing army worm insect detection and classification model using deep convolution neural network. https://ir.bdu.edu.et/handle/123456789/10890?show=full
- Borgli, H., Vajira Thambawita, V., Smedsrud, P. H., Hicks, S., Jha, D., Sigrun L. Eskeland,S. L., Randel, K. R., Pogorelov, K., Lux, M., Nguyen,D. T. D., Johansen, D., Griwodz, C., Stensland, H. K., Garcia-Ceja, E., Schmidt, P. T., Hammer, H. L., Riegler, M. A., Halvorsen, P., Thomas de Lange. (2020). HyperKvasir, a comprehensive multi-class image and video dataset for gastrointestinal endoscopy. Scientific Data, 7(1), 1–14. https://doi.org/10.1038/s41597-020-00622-y
- Chollet, F. (2017). Xception: Deep learning with depthwise separable convolutions. Proceedings - 30th IEEE conference on computer vision and pattern recognition, CVPR 2017, 2017 January, 1800–1807. https://doi.org/10.1109/CVPR.2017.195
- Deng, J., Dong, W., Socher, R., Li, L., Li, K., & Fei-fei, L. (2009). ImageNet : A Large-scale hierarchical image database. IEEE Conference on Computer Vision and Pattern Recognition (pp. 248–255). https://doi.org/10.1109/CVPR.2009.5206848.
- Escobar, J., Sanchez, K., Hinojosa, C., Arguello, H., & Castillo, S. (2021). Accurate deep learning-based gastrointestinal disease classification via transfer learning strategy. XXIII Symposium on Image, Signal Processing and Artificial Vision (STSIVA), 1–5. https://doi.org/10.1109/STSIVA53688.2021.9591995.
- Gamage, C., Wijesinghe, I., Chitraranjan, C., & Perera, I. (2019). GI-Net: Anomalies classification in gastrointestinal tract through endoscopic imagery with deep learning,” MERCon 2019 - Proceedings, 5th International Multidisciplinary Moratuwa Engineering Research Conference, 66–71, https://doi.org/10.1109/MERCon.2019.8818929
- Guan, F., Ton, P., Ge, S., & Zhao, L. (2014). Anisotropic diffusion filtering for ultrasound speckle reduction. Science China Technological Sciences, 57(3), 607–614. https://doi.org/10.1007/s11431-014-5483-7
- Guo, Y., Liu, Y., Oerlemans, A., Lao, S., Wu, S., & Lew, M. S. (2016). Deep learning for visual understanding: A review. Neurocomputing, 187, 27–48. https://doi.org/10.1016/j.neucom.2015.09.116
- Hirasawa, T., Aoyama, K., Tanimoto, T., Ishihara, S., Shichijo, S., Ozawa, T., Ohnishi, T., Fujishiro, M., Matsuo, K., Fujisaki, J., & Tada, T. (2018). Application of artificial intelligence using a convolutional neural network for detecting gastric cancer in endoscopic images. Gastric Cancer: Official Journal of the International Gastric Cancer Association and the Japanese Gastric Cancer Association, 21(4), 653–660. https://doi.org/10.1007/s10120-018-0793-2
- Jain, S., & Salau, A. O. (2019). An image feature selection approach for dimensionality reduction based on kNN and SVM for AKT Proteins. Cogent Engineering, 6(1), 1–14. https://doi.org/10.1080/23311916.2019.1599537
- Jha, D., Ali, S., Hicks, S., Thambawita, V., Borgli, H., Smedsrud, H. P., & Halvorsen, P. (2021). A comprehensive analysis of classification methods in gastrointestinal endoscopy imaging. Medical Image Analysis, 70(1), 102007. https://doi.org/10.1016/j.media.2021.102007
- Karargyris, A., & Bourbakis, N. (2011). Detection of small bowel polyps and ulcers in wireless capsule endoscopy videos. IEEE Transactions on Biomedical Engineering, 58(10 PART 1), 2777–2786. https://doi.org/10.1109/TBME.2011.2155064
- Khan, A., Sohail, A., Zahoora, U., & Qureshi, A. S. (2020). A survey of the recent architectures of deep convolutional neural networks. Artificial Intelligence Review, 53(8), 5455–5516. https://doi.org/10.1007/s10462-020-09825-6
- Khan, M. A., Khan, M. A., Ahmed, F., Mittal, M., Goyal, L. M., Jude Hemanth, D., & Satapathy, S. C. (2020). Gastrointestinal diseases segmentation and classification based on duo-deep architectures. Pattern Recognition Letters, 131(1), 193–204. https://doi.org/10.1016/j.patrec.2019.12.024
- Khan, M. A., Sarfraz, M. S., Alhaisoni, M., Albesher, A. A., Wang, S., & Ashraf, I. (2020). StomachNet: Optimal deep learning features fusion for stomach abnormalities classification. IEEE Access, 8(1), 197969–197981. https://doi.org/10.1109/ACCESS.2020.3034217
- Khan, M. A., Majid, A., Hussain, N., Alhaisoni, M., Zhang, Y., Kadry, S., & Nam, Y. (2021). Multiclass stomach diseases classification using deep learning features optimization. Computers, Materials & Continua, 67(3), 3382–3398. https://doi.org/10.32604/cmc.2021.014983
- Khan, M. A., Nasir, I. M., Sharif, M., Alhaisoni, M., Kadry, S., Bukhari, S. A. C., & Nam, Y. (2021). A blockchain based framework for stomach abnormalities recognition. Computers, Materials & Continua, 67(1), 141–158. https://doi.org/10.32604/cmc.2021.013217
- Li, B., & Meng, M. Q. (2012). Expert systems with applications automatic polyp detection for wireless capsule endoscopy images. Expert Systems With Applications, 39(12), 10952–10958. https://doi.org/10.1016/j.eswa.2012.03.029
- Li, B., & Meng, M. Q. H. (2012). Tumor recognition in wireless capsule endoscopy images using textural features and SVM-based feature selection. IEEE Transactions on Information Technology in Biomedicine, 16(3), 323–329. https://doi.org/10.1109/TITB.2012.2185807
- Li, C., Zhang, S., Zhang, H., Pang, L., Lam, K., Hui, C., & Zhang, S. (2012). Using the K-Nearest neighbor algorithm for the classification of lymph node metastasis in gastric cancer. Computational and Mathematical Methods in Medicine, 2012, 1–11. https://doi.org/10.1155/2012/876545
- Lin, W., Hasenstab, K., Cunha, G. M., Schwartzman, A., & Elbe-Bürger, A. (2020). Comparison of handcrafted features and convolutional neural networks for liver MR image adequacy assessment. Scientific Reports, 10(1), 1–11. https://doi.org/10.1038/s41598-020-77264-y
- Lonseko, Z. M., Adjei, P. E., Du, W., Luo, C., Hu, D., Zhu, L., Gan, T., & Rao, N. (2021). Gastrointestinal disease classification in endoscopic images using attention-guided convolutional neural networks. Applied Sciences, 11(23), 11136. https://doi.org/10.3390/app112311136
- Maier-hein, L., Vedula, S. S., Speidel, S., Navab, N., Kikinis, R., Park, A., Eisenmann, M., Feussner, H., Forestier, G., Giannarou, S., Hashizume, M., Katic, D., Kenngott, H., Kranzfelder, M., Malpani, A., März, K., Neumuth, T., Padoy, N., Pugh, C., Schoch, N., Stoyanov, D., Taylor, R., Wagner, M., Hager, D. G., & Jannin, P. (2017). Surgical data science for next- generation interventions. Nature Biomedical Engineering, 1(9), 691–696. https://doi.org/10.1038/s41551-017-0132-7
- Majid, A., Khan, M. A., Yasmin, M., Rehman, A., Yousafzai, A., & Tariq, U. (2020). Classification of stomach infections: A paradigm of convolutional neural network along with classical features fusion and selection. Microscopy Research and Technique, 83(5), 562–576. https://doi.org/10.1002/jemt.23447
- Mengistie, Y. (2020). The pattern and outcome of upper gastrointestinal bleeding at St. Paul's Millenium Medical College, Addis Ababa, Ethiopia. Ethiopian Medical Journal, 58(4), 323–327.
- Naz, J., Sharif, M., Yasmin, M., Raza, M., & Khan, M. A. (2021). Detection and classification of gastrointestinal diseases using machine learning. Curr Med Imaging, 17(4), 479–490. https://doi.org/10.2174/1573405616666200928144626
- Noor Mohamed, S. S., & Srinivasan, K. (2020). Comparative analysis of deep neural networks for crack image classification. In Lecture Notes on Data Engineering and Communications Technologies (Vol. 38, pp. 434–443). Springer. https://doi.org/10.1007/978-3-030-34080-3_49
- Owais, M., Arsalan, M., Choi, J., Mahmood, T., & Park, K. R. (2019). Artificial intelligence-based classification of multiple gastrointestinal diseases using endoscopy videos for clinical diagnosis. Journal of Clinical Medicine, 8(7), 986. https://doi.org/10.3390/jcm8070986
- Öztürk, S., & Özkaya, U. (2020). Residual LSTM layered CNN for classification of gastrointestinal tract diseases. Journal of Biomedical Informatics, 103638, 1–31. https://doi.org/10.1016/j.jbi.2020.103638
- Paoletti, M. E., Haut, J. M., Tao, X., Miguel, J. P., & Plaza, A. (2020). A new GPU implementation of support vector machines for fast hyperspectral image classification. Remote Sensing, 12(8), 1257. https://doi.org/10.3390/RS12081257
- Ramzan, M., Raza, M., Sharif, M., Khan, M. A., & Nam, Y. (2021). Gastrointestinal tract infections classification using deep learning. Computers, Materials & Continua, 67(3), 3239–3257. https://doi.org/10.32604/cmc.2021.015920
- Sakib, S., Nazib, A., Jawad, A., Kabir, J., & Ahmed, H. (2018). An overview of convolutional neural network. Its Architecture and Applications. https://doi.org/10.20944/preprints201811.0546.v1
- Salau, A. O., & Jain, S. (2019). Feature extraction: A survey of the types, techniques, and applications. 5th IEEE International Conference on Signal Processing and Communication (ICSC), Noida, India, IEEE, 158–164. https://doi.org/10.1109/ICSC45622.2019.8938371
- Salau, A. O., & Jain, S. (2021). Adaptive Diagnostic machine learning technique for classification of cell decisions for AKT protein. Informatics in Medicine Unlocked, 23(1), 1–9. https://doi.org/10.1016/j.imu.2021.100511
- Salau, A. O. (2021). Detection of corona virus disease using a novel machine learning approach. International Conference on Decision Aid Sciences and Application (DASA), 587–590. https://doi.org/10.1109/DASA53625.2021.9682267
- Segu, S., Drozdzal, M., Pascual, G., Radeva, P., Malagelada, C., Azpiroz, F., & Vitrià, J. (2016). Generic feature learning for wireless capsule endoscopy analysis. Computers in Biology and Medicine, 79(1), 163–172. https://doi.org/10.1016/j.compbiomed.2016.10
- Sharif, M., Attique Khan, M., Rashid, M., Yasmin, M., Afza, F., & Tanik, U. J. (2019). Deep CNN and geometric features-based gastrointestinal tract diseases detection and classification from wireless capsule endoscopy images. Journal of Experimental and Theoretical Artificial Intelligence, 1–23. https://doi.org/10.1080/0952813X.2019.1572657
- Shetty, S. K., & Siddiqa, A. (2019). Deep learning algorithms and applications in computer vision. International Journal of Computer Sciences and Engineering, 7(7), 195–201. https://doi.org/10.26438/ijcse/v7i7.195201
- Shichijo, S., Nomura, S., Aoyama, K., Nishikawa, Y., Miura, M., Shinagawa, T., Takiyama, H., Tanimoto, T., Ishihara, S., Matsuo, K., & Tada, T. (2017). EBiomedicine application of convolutional neural networks in the diagnosis of Helicobacter pylori infection based on endoscopic images. EBioMedicine, 25(1), 106–111. https://doi.org/10.1016/j.ebiom.2017.10.014
- Shorten, C., & Khoshgoftaar, T. M. (2019). A survey on image data augmentation for deep learning. Journal of Big Data, 6(1), 1–48. https://doi.org/10.1186/s40537-019-0197-0
- Simonyan, K., & Zisserman, A. (2015). Very deep convolutional networks for large-scale image recognition. In 3rd International Conference on Learning Representations, ICLR 2015 - Conference Track Proceedings, 1–14.
- Souaidi, M., Ait, A., & El Ansari, M. (2019). Multi-scale completed local binary patterns for ulcer detection in wireless capsule endoscopy images. Multimedia Tools and Applications, 78(10), 13091–13108. https://doi.org/10.1007/s11042-018-6086-2
- Szegedy, C., Liu, W., Jia, Y., Sermanet, P., Reed, S., Anguelov, D., Erhan, D., Vanhoucke, V., Rabinovich, A. (2015). Going deeper with convolutions. Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, 07–12June, 1–9. https://doi.org/10.1109/CVPR.2015.7298594
- Takiyama, H., Ozawa, T., Ishihara, S., Fujishiro, M., Shichijo, S., Nomura, S., Miura, M., & Tada, T. (2018). Automatic anatomical classification of esophagogastrodenoscopy images using deep convolutional neural networks. Scientific Reports, 8(1), 1–8. https://doi.org/10.1038/s41598-018-25842-6
- Wang, Y., Tavanapong, W., Wong, J., Hwan, J., & De Groen, P. C. (2015). Polyp-Alert : Near real-time feedback during colonoscopy. Computer Methods and Programs in Biomedicine, 120(3), 164–179. https://doi.org/10.1016/j.cmpb.2015.04.002
- Woreta, S. A., Yassin, M. O., Teklie, S. Y., Getahun, G. M., & Abubeker, Z. A. (2015). Upper gastrointestinal endoscopy findings at Gondar university international journal of pharmaceuticals and health care research. International Journal of Pharmaceuticals and Health Care Research, 3(2), 60–65.
- Yuan, Y., & Meng, M. Q. (2014). Polyp classification based on bag of features and saliency in wireless capsule endoscopy. IEEE International Conference on Robotics and Automation (ICRA), 3930–3935.