Abstract
Maize chlorotic mottle virus (MCMV) infects maize plants and causes significant losses in corn production worldwide. In this study, a real-time TaqMan RT-PCR assay for efficient detection of MCMV was described. A pair of primers amplifying a 131-bp DNA fragment and a TaqMan probe was designed targeting the novel molecular marker based on MCMV genome analysis sequences. The assay designed was highly specific, producing no signal from other viruses, and the sensitivity of the assay was 0.16 fg/reaction of total RNA, which was approximately 252-fold higher than conventional RT-PCR gel electrophoresis method. Compared with the real-time TaqMan reverse transcription PCR targeting coat protein gene, the novel assay has more specificity and sensitivity to detect MCMV in maize. Therefore, the assay is a useful tool for large or middle-scale corporations and entry-exit inspection and quarantine bureau to detect MCMV in maize seeds or plant tissues.
Public Interest Statement
Maize chlorotic mottle virus (MCMV) infects maize plants and causes significant losses in corn production worldwide, and it has been a quarantine pest by the Chinese government in 2007. Many kinds of assays have been reported for detection of MCMV, such as biological indexing, enzyme-linked immunosorbent assay (ELISA), electron microscopy, surface plasmon resonanceand biosensor based on a quartz crystal microbalance. However, these assays have some disadvantages. Simultaneously, coat protein gene is not specific to MCMV enough. Therefore, there is an urgent need for mining a novel molecular marker specific to MCMV and developing a real-time RT-PCR for detection of MCMV. In our study, a real-time TaqMan RT-PCR assay for efficient detection of MCMV was developed with a novel molecular marker according to microbial genome analysis. The novel assay is a real good performance than ever.
Competing Interests
The authors declare no competing interest.
1. Introduction
Maize chlorotic mottle virus (MCMV) is only species in the genus Machlomovirus (family Tombusviridae), and it is the sense single strand RNA virus. MCMV was an important plant pathogenic virus, infected Zea mays in Peru (Castillo & Hebert, Citation1974) and lead to losses of 10–15% in floury and sweet corn cultivars. Furthermore, once MCMV the combines with the Maize dwarf mosaic virus, Sugarcane mosaic virus, or Wheat streak mosaic virus, it may induce the severe disease known as maize lethal necrosis (Wu et al., Citation2013). In addition, this virus can be carried readily into other countries or regions by seeds or vectors (Cabanas & Bressan, Citation2011; Cabanas, Watanabe, Higashi, & Bressan, Citation2013). It had the potential threat to the production of maize crops, so it became a quarantine pest by the Chinese government in 2007 (Wen, Zhang, Cui, & Zhang, Citation2011), and was identified in maize seeds imported from the United States (Niblett & Claflin, Citation1978), Congo (Lukanda et al., Citation2014), and Kenya (Kusia et al., Citation2015). A high risk of MCMV has been introduced with the increasing international exchange of maize seeds. In recent year, severe chlorotic mottle symptoms in sweet corns or sugarcanes were observed in leaves infected with MCMV in many countries and regions (Deng, Chou, Chen, Tsai, & Lin, Citation2014; Lukanda et al., Citation2014; Wang, Zhou, & Wu, Citation2014). In order to prevent introduction of MCMV through international exchange of maize seeds and MCMV transmission, there is an urgent need for a reliable and sensitive assay for detection of MCMV. Many kinds of assays have been reported for detection of MCMV, such as biological indexing (Uyemoto, Citation1983), ELISA (Wu et al., Citation2013), electron microscopy (Morales, Arroyave, Castillo, & De Leon, Citation1999), surface plasmon resonance (Zeng et al., Citation2013) and biosensor based on a quartz crystal microbalance (Huang, Xu, Ji, Li, & Chen, Citation2014). However, these assays have many disadvantages. Biological indexing is time-consuming, labor-intensive and requires greenhouse space; The results of ELISA are dependent on the quality and availability of expensive antibodies; Electron microscopy is laborious and time-consuming, simultaneously it also requires expensive instruments and virology knowledge (Zhang, Hung, Song, & He, Citation2013); Surface plasmon resonances require very expensive equipment.
Real-time reverse transcription PCR (RT-PCR) is an innovative evolution of conventional reverse transcript PCR, which has been used as a major analytical platform in molecular biology. The incorporation of fluorescence-based detection systems into real-time PCR instruments deal with the kinetic detection of the accumulation of PCR products over the cycling period and greater sensitivity to amplicon detection than conventional gel-based detection. Because of its improved rapidity, sensitivity, reproducibility, reduced risk of carry-over contamination, and ability to quantify viral nucleic acid directly from samples, real-time PCR assays have utilized widely than conventional PCR assays in the diagnosis of viral diseases (Callison et al., Citation2007).
To date, nine complete genome sequences representing the isolates of MCMV have been made available in NCBI website. In this study, a real-time TaqMan RT-PCR assay by targeting a novel detection marker based on complete genome sequences analysis was currently developed. The specificity and sensitivity of real-time PCR protocol were determined and this assay was applied for detection of artificial samples. This research served as a foundation for further application of MCMV detection in the wide fields.
2. Material and methods
2.1. Materials and reagents
Six MCMV and seven other virus isolates were employed in this study. MCMV-ZJ and MCMV-YP were supplied from Beijing Entry-Exit Inspection and Quarantine Bureau, Beijing, China; MCMV-BJ was obtained from China Agricultural University; MCMV-agdia2094 was purchased from Agdia, USA; MCMV-ATCC-PV262 were purchased from American Type Culture Collection (Manassas, USA); MCMV-1087 isolate was obtained from Braunschweig, Germany; Maize dwarf mosaic virus (MDMV), Arabis mosaic virus (ArMV), Carnation ringspot virus (CRSV), Odontoglossum ringspot virus (ORSV), Cucumber green mottle mosaic virus (CGMMV), Lily symptomless virus (LSV), Tobacco rattle virus(TRV) isolates were kept in Shanghai Entry-Exit Inspection and Quarantine Bureau, Shanghai, China. MCMV and MDMV viruses were inoculated in Zea mays, and the infected tissues were harvested after 15 days. ArMV, CRSV and TRV were inoculated in Chenopodium quino, CGMMV were inoculated in Cucumissativus, and the infected tissues were harvested after 14 days. The leaf of Phalaenopsis aphrodite with ORSV, and the lily bulbs with LSV were intercepted in entry plants in Shanghai Entry-Exit Inspection and Quarantine Bureau.
2.2. Viral RNA extraction
Viral genomic RNA was extracted from 100 mg of leaves infected with the viruses using TIANamp Virus RNA Kit (Tiangen Biotechnology, Beijing, China) according to the manufacturer’s protocols. The RNA was eluted with 80 μl of elution buffer and stored at −20°C.
2.3. Primers and probe design
A sequence alignment of all of nine MCMV genomes (GenBank accession No. KP851970.1, NC_003627.1, KJ782300.1, KF010583.1, JQ982470.1, JQ982469.1, GU138674.1, EU358605.1 and X14736.2) was used to identify conserved sequence to design primers and a probe for the real-time RT-PCR assay. A forward primer (FP), a reverse primer RP, and a Taqman® dual-labeled probe were listed in Table . The specificity of the designed primers was examined using the Primer BLAST online alignments at the website (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). The primers and probes were synthesized by Shanghai Gencore Biotechnologies, Co, Ltd (Shanghai, China).
Table 1. Primes and probe used in the study
2.4. RT-PCR and real-time TaqMan RT-PCR
RT-PCR assays were performed on Applied Biosystems® 2720 Thermal Cycler (Life Technologies, MA, USA) using One-Step PrimeScriptTM RT-PCR Kit (Takara, China). All sets of reactions were carried out in a final volume of 25 μl, which consisted of 0.50 μl of RNA (MCMV), 12.50 μl of 2 × OneStep RT-PCR Buffer, 1 μl of PrimeScript enzyme (5U/μl), 0.50 μl of MCMV FP (20 μM), 0.50 μl of MCMV RP (20 μM), and 10 μl of RNase-free ddH2O. Amplification reactions were made up of a reverse transcription at 50°C for 30 min, an initial denaturing step at 94°C for 3 min followed by 35 cycles at 94°C for 30s, 58°C for 30s and 72°C for 30s followed by a final extension at 72°C for 10 min. The amplicons were analyzed by electrophoresis through a 1.50% agarose gel and stained with ethidium bromide.
Twenty-five μl of real-time TaqMan RT-PCR reactions were prepared with 2 μl of RNA, 12.50 μl of 2 × buffer, 0.20 μl of Ex Taq HS (5 μM), 0.20 μl of PrimeScript RT enzyme MIXⅡ (Takara, China),0.20 μl of MCMV FP (20 μM), 0.20 μl of MCMV RP (20 μM), 0.40 μl of probe (20 μM), 0.20 μl of ROX Reference DyeⅡ (50 × ) and 9.10 μl of RNase-free ddH2O. Amplification reactions were carried out with the following program: incubation for 30 min at 50°C and then predenature at 95°C for 5 min, followed by 40 cycles of 95°C for 10s and 60°C for 1 min. Real-time TaqMan RT-PCR were carried out on ViiA™ 7 Real-Time PCR apparatuses (Life Technologies, MA, USA) using One-Step PrimeScriptTM RT-PCR Kit.
2.5. Specificity and sensitivity of real-time TaqMan PCR
The specificity of this TaqMan assay was evaluated by comparing MCMV with ATCC-PV262, MDMV, ArMV, CRSV, ORSV, CGMMV, LSV, and TRV. In addition, fluorescent signals were recorded simultaneously.
The sensitivities of the real-time TaqMan assay and conventional PCR were determined using a 10-fold serial dilution of RNA between 80.70 pg/μl and 0.81 fg/μl of MCMV-ATCC-PV262. Sensitivity was the lowest concentration of real-time RT-PCR.
2.6. Validation of real-time TaqMan RT-PCR assay with artificial infected leaves
The performance of this assay was studied using the total RNA of sole maize leaves infected with MCMV or other leaves infected with non-MCMV. Total RNA extract and real-time TaqMan PCR were carried out according to previous protocols.
3. Results
3.1. Primers and probe
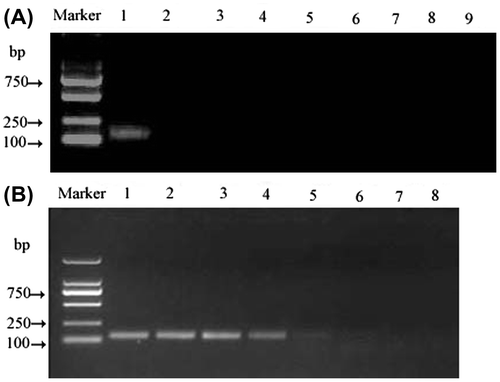
After nine complete genomes of MCMV were aligned, alignment report showed that the region from nucleotides 4,150 to 4,280 had only three different nucleotides, which indicated a very high species-conserved to MCMV (Figure ). Furthermore, the conserved sequence was aligned with other genomes data using BLAST, the conserved sequence had the over 98% identity to other uncompleted genomes of MCMV with E values less than 7e−59; therefore, it could be regarded as a molecular marker. Based on the conserved sequence, the specific primers and probe were designed and synthesized, the products of conventional RT-PCR with primers (FP and RP), using One-Step PrimeScript TM RT-PCR Kit (Takara, China) were verified by electrophoresing on a 1.50% agarose gel (Figure (A)). The result showed that the RT-PCR produced an intense band with the expected 131 bp for MCMV.
Figure 1. Alignment of the TaqMan real-time RT-PCR amplified region. All of nine MCMV isolates with complete genome were included. Forward and reverse primers and the probe target site are indicated.
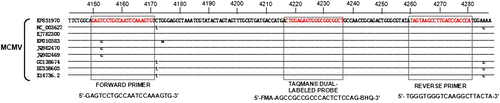
Figure 2. RT-PCR for MCMV detection by agarose gel electrophoresis. (A) The specificity of RT-PCR. Lane M: DL2000 DNA marker; lane 1: MCMV-ATCC-PV262; lane 2: MDMV, lane 3: ArMV, lane 4: CRSV, lane 5: ORSV, lane 6: CGMMV, lane 7: LSV, lane 8: TRV and lane 9: distilled water. (B) The sensitivity of RT-PCR for detection of MCMV-ATCC-PV262. Marker: DL2000 DNA marker(Takara, China); lane 1: 40.35 pg/reaction; lane 2: 4.04 pg/reaction; lane 3: 404 fg/reaction; lane 4: 40.40 fg/reaction; lane 5: 4.04 fg/reaction; lane 6: 0.404 fg/reaction; lane 7: 0.04 fg/reaction; lane 8: negative control with no template RNA.
3.2. Specificity of real-time TaqMan PCR
Based on the TaqMan probe, strong fluorescent signals were detected only from reactions with MCMV, while no signals were detected for non-MCMV viruses along with the water control were determined (Figure (B)), which clearly described that MCMV could be clearly differentiated from other viruses by comparing the signals. The PCR products were analyzed further by agarose gel electrophoresis. The assay with MCMV sample displayed the expected band of 131 bp (Figure (A), lane 1), whereas those negative did not happen. These results suggested that this probe and the primers had specificity for the MCMV.
3.3. Sensitivity comparison of real-time RT-PCR and conventional RT-PCR
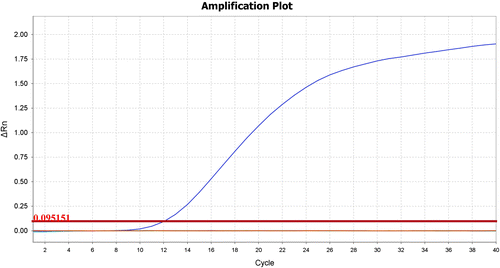
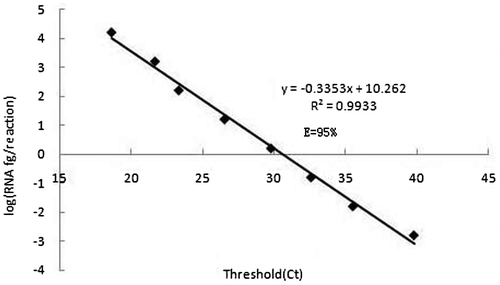
There were apparent specific amplification products with MCMV RNA as templates from 40.35 pg/reaction to 4.04 fg/reaction, and there also existed faint specific band in lane 5 with the 4.04 fg/reaction of RNA (Figure ). However, it could not be regarded as the lowest determination limit because of a faint band, the lowest detection limit of the conventional RT-PCR was 40.35 fg/reaction of RNA. In real-time TaqMan RT-PCR assay, Figure indicated that all of specific amplifications were carried out with RNA templates from 161.40 pg/reaction to 0.16 fg/reaction with Ct less than 36, and the lowest detection limit was 0.16 fg/reaction of RNA. Therefore, real-time TaqMan RT-PCR was about 252 (40.35/0.16) times more sensitive than conventional RT-PCR.
3.4. The performance of the real-time TaqMan PCR
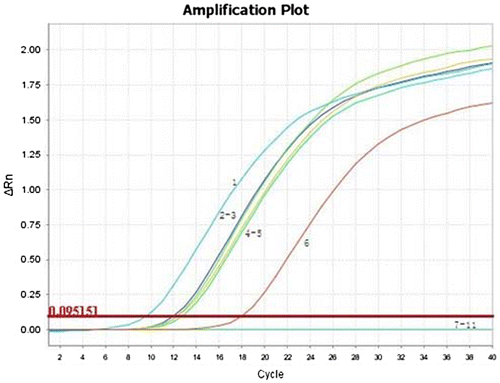
The validation tests indicated that all six maize leaves infected MCMV showed positive results, and their Ct values were 9.53, 12.00, 12.30, 12.35, 12.76, 17.97, respectively, whereas no Ct values were detected with other leaves infected with non-MCMV viruses and negative controls by this assay (Figures and ). This result confirmed further the reliability of this TaqMan RT-PCR assay for routine testing.
Figure 4. Standard curves of the developed TaqMan real-time RT-PCR assay using the specific probe. The linear dynamic range was established among 160.14 pg/reaction, 16.01 pg/reaction, 1.60 pg/reaction, 160 fg/reaction, 16.0 fg/reaction, 1.60 fg/reaction 0.16 fg/reaction and 0.02 fg/reaction RNA of MCMV. Each point represents the mean Ct (eight independent reactions, three replicates each). The coefficient of determination (R2) and the efficiency (E) of each linear regression curve is indicated.
4. Discussion
Since MCMV is a mandatory quarantine pathogen in China, accurate identification and detection of this virus play a crucial role in successful containment of its introduction or further spread. A rapid, sensitive, real-time RT-PCR assay based on TaqMan chemistry was set up for efficient detection of MCMV in tubes, in addition to the conventional RT-PCR protocol in our study. Although real-time RT-PCR technique was more sensitive (252-folds) compared with conventional RT-PCR assay, the latter method may be preferred as an alternative to the real-time technique in some laboratories owing to nonavailabilty of sophisticated equipment. These two methods may assist in enforcing quarantine regulation by reliable detection of MCMV and in routine indexing of MCMV for production of virus-free maize areas.
Molecular detection markers play an important role in any microorganism for species discrimination, and promote developments of molecular detection assays. Despite the fact that coat protein genes are popular to be used as detection markers for various viruses (Barthe, Ceccardi, Manjunath, & Derrick, Citation1998; Bennani, Mendes, Zemzami, Azeddoug, & Nolasco, Citation2002; Bhat, Bhadramurthy, Siju, & Hareesh, Citation2006; Good & Monis, Citation2001), there are different fragments to be used as makers for virus detection (Callison et al., Citation2006; Sane et al., Citation2012; Yue et al., Citation2008). Recently, with increasing development of genome sequencing, 81106 microbial species have been completely sequenced (Ncbi 2015), therefore, mining the species-specific detection marker has been available in our previous study using large-scale genome data (Hang, Liu, Gao, & Yin, Citation2009; Liu, Shi, & Pan, Citation2007; Liu et al., Citation2015). Based on the similar strategy, candidate molecular detection marker had been investigated and located at the position of 4,000–4,437 of MCMV isolate Nebraska genome. The novel molecular detection marker has high identity over 98% of MCMV than others throughout blast analysis, simultaneously; there exists three 100% identity, fifteen 99% identity, and four 98% identity. However, coat protein gene (711 bp) has the identity of over 96% to MCMV than others throughout blast analysis, and only one 100% identity appears, twice 96% identity happens. Therefore, the novel molecular detection marker is more species-specific to MCMV than coat protein according to our bioinformatics analysis. In addition, RT-PCR can amplify the specific products of 131 bp with MCMV samples, and non-MCMV samples do not show the specific band in all of samples, which indicate that the novel molecular detection marker has the good specificity in biological experiments.
In a real-time PCR assay a positive reaction is detected by accumulation of a fluorescent signal. The Ct (cycle threshold) is described as the number of cycles required for the fluorescent signal to cross the threshold. Ct values are inversely proportional to the amount of target nucleic acid in the sample, the lower the Ct value, the greater the amount of target nucleic acid in the samples. In general, Ct values less than 29 are strong positive reactions indicative of abundant target nucleic acid in the sample, and Ct values of 30–37 are positive reactions indicative of moderate amounts of target nucleic acid. Zhang et al. developed real-time TaqMan RT-PCR detection methods targeting coat protein gene, and the result showed low sensitivity for detection of MCMV in maize seeds is 4 fg/reaction of RNA with a Ct value of 36.20 (Zhang et al., Citation2011), However, in our study the sensitivity of real-time TaqMan RT-PCR for detection of MCMV in maize leaves based on the novel detection marker is 0.16 fg/reaction of total RNA with a Ct value of 35.546, it demonstrates that the novel real-time TaqMan RT-PCR in our experiment has higher (25 folds) sensitivity than other detection markers. From the biological experiment and bioinformatics analysis, the present study developed a rapid, species-specific real-time TaqMan RT-PCR assay for MCMV detection. Our study indicates that it is available for researchers to mine species-specific markers for detection of microbial containing bacteria and viruses with large-scale microbial genomes bioinformatics analysis. It is beneficial for increasing novel molecular markers for detection of pathogen.
China has strengthened the prevention and control of introduction of maize seed through international exchange of maize seeds, there is seldom report MCMV transitions, therefore, it is very difficult to gain field isolates. In order to evaluate the novel assay for MCMV detection, MCMV transmission in the corn field was simulated in our lab, and artificially infected samples were prepared. The results showed that the positive results of infected samples were the same, which was due to the specificity and reliability of the real-time RT-PCR. This assay only required simple sample preparation and the results were obtained in a few hours, which make it a promising assay. Since we did not have access to field isolates, the current method has only been tested on the limited number of known MCMV isolate. Therefore, we hope to be able to obtain more trials with field isolated viruses to tamp fundamentals for assay applications.
Sugarcane and corn in field were also found to be infected with MCMV in Yunnan Province, China in 2013. The method for RNA extracting from plant tissues is a conventional protocol, therefore the novel assay for detection of MCMV is very suitable for the detection of field leaves samples without than other assays according to only our sample preparation and RT-PCR protocol.
In this work, we developed a specific real-time Taq-Man RT-PCR assay. The assay described is high specificity, sensitivity, and can be used for the rapid detection of MCMV.
Additional information
Funding
Notes on contributors
Cuiyun Yang
Professor Cuiyun Yang is working as a Young Scientist (Principal Investigator) in the Shanghai Entry-Exit Inspection and Quarantine Bureau, Shanghai, China. Her area of research is to develop the standard of detection of plant pathogen. She has intercepted decades of quarantine pests by the Chinese government. Currently she has been engaged with two Science Projects of General Administration of Quality Supervision, Inspection and Quarantine of the People's Republic of China, eight Science Projects of Committee of Science and Technology of Shanghai, and developed eight national standards and four industry standards on pathogen detection. She has published more sixty research articles, three books, and gained four authorized patents, five Provincial Awards, Science and Technology Progress Award of Shanghai Pudong New Area.
References
- Barthe, G. A., Ceccardi, T. L., Manjunath, K. L., & Derrick, K. S. (1998). Citrus psorosis virus: nucleotide sequencing of the coat protein gene and detection by hybridization and RT-PCR. Journal of General Virology, 79, 1531–1537.10.1099/0022-1317-79-6-1531
- Bennani, B., Mendes, C., Zemzami, M., Azeddoug, H., & Nolasco, G. (2002). Citrus variegation virus: Molecular variability of a portion of the RNA 3 containing the coat protein gene and design of primers for RT-PCR detection. European Journal of Plant Pathology, 108, 155–162.10.1023/A:1015009019552
- Bhat, A. I., Bhadramurthy, V., Siju, S., & Hareesh, P. S. (2006). Detection and identification of cymbidium mosaic virus infecting vanilla (Vanilla planifolia Andrews) in India based on coat protein gene sequence relationships. Journal of Plant Biochemistry and Biotechnology, 15, 33–37.10.1007/BF03321898
- Cabanas, D., & Bressan, A. (2011). Dissecting the mode of transmission of Maize chlorotic mottle virus by the corn thrips, Frankliniella williamsi. Phytopathology, 101, S24–S24.
- Cabanas, D., Watanabe, S., Higashi, C. H. V., & Bressan, A. (2013). Dissecting the mode of maize chlorotic mottle virus transmission (Tombusviridae: Machlomovirus) by Frankliniella williamsi (Thysanoptera: Thripidae). Journal of Economic Entomology, 106, 16–24.10.1603/EC12056
- Callison, S. A., Hilt, D. A., Boynton, T. O., Sample, B. F., Robison, R., Swayne, D. E., & Jackwood, M. W. (2006). Development and evaluation of a real-time taqman Rt-PCR assay for the detection of infectious Bronchitis virus from infected chickens. Journal of Virological Methods, 138, 60–65.10.1016/j.jviromet.2006.07.018
- Callison, S. A., Riblet, S. M., Oldoni, I., Sun, S., Zavala, G., Williams, S. … García, M. (2007). Development and validation of a real-time taqman® PCR assay for the detection and quantitation of infectious Laryngotracheitis virus in poultry. Journal of Virological Methods, 139, 31–38.10.1016/j.jviromet.2006.09.001
- Castillo, J., & Hebert, T. T. (1974). A new virus disease of maize in Peru. Fitopatologia, 9, 79–84.
- Deng, T. C., Chou, C. M., Chen, C. T., Tsai, C. H., & Lin, F. C. (2014). First report of Maize chlorotic mottle virus on sweet corn in Taiwan. Plant Disease, 98, 1748.
- Good, X., & Monis, J. (2001). Partial genome organization, identification of the coat protein gene, and detection of grapevine leafroll-associated virus-5. Phytopathology, 91, 274–281.10.1094/PHYTO.2001.91.3.274
- Hang, B., Liu, Z. M., Gao, H. Y., & Yin, J. Y. (2009). Data mining for molecular detection markers of Listeria monocytogenes. Food Science [in Chinese], 197–200.
- Huang, X., Xu, J. M., Ji, H. F., Li, G. F., & Chen, H. J. (2014). Quartz crystal microbalance based biosensor for rapid and sensitive detection of Maize chlorotic mottle virus. Anal Methods-UK, 6, 4530–4536.
- Kusia, E. S., Subramanian, S., Nyasani, J. O., Khamis, F., Villinger, J., Ateka, E. M., & Pappu, H. R. (2015). First report of lethal necrosis disease associated with co-infection of finger millet with Maize chlorotic mottle virus and sugarcane mosaic virus in kenya. Plant Disease, 99, 899–900.10.1094/PDIS-10-14-1048-PDN
- Liu, Z. M., Shi, X. M., & Pan, F. (2007). Species-specific diagnostic marker for rapid identification of Staphylococcus aureus. Diagnostic Microbiology and Infectious Disease, 59, 379–382.10.1016/j.diagmicrobio.2007.06.011
- Liu, Z. M., Zhu, J. C., Xia, X. Y., Wang, L., Yang, C. Y., & Li, X. H. (2015). Development of a loop-mediated isothermal amplification assay based on lmo0460 sequence for detection of Listeria monocytogenes. Journal of Food Safety, 35, 362–369.10.1111/jfs.2015.35.issue-3
- Lukanda, M., Owati, A., Ogunsanya, P., Valimunzigha, K., Katsongo, K., Ndemere, H., & Kumar, P. L. (2014). First report of Maize chlorotic mottle virus infecting maize in the Democratic Republic of the Congo. Plant Disease, 98, 1448–1448.10.1094/PDIS-05-14-0484-PDN
- Morales, F. J., Arroyave, J. A., Castillo, J., & De Leon, C. (1999). Cytopathology of Maize chlorotic mottle virus in Zea mays L. Maydica, 44, 231–235.
- Niblett, C. L., & Claflin, L. E. (1978). Corn lethal necrosis-a new virus disease of corn in Kansas. Plant Disease Reporter, 62, 15–19.
- Sane, J., Kurkela, S., Levanov, L., Nikkari, S., Vaheri, A., & Vapalahti, O. (2012). Development and evaluation of a real-time RT-PCR assay for Sindbis virus detection. Journal of Virological Methods, 179, 185–188.10.1016/j.jviromet.2011.10.021
- Uyemoto, J. K. (1983). Biology and control of Maize chlorotic mottle virus. Plant Disease, 67, 7–10.10.1094/PD-67-7
- Wang, Q., Zhou, X. P., & Wu, J. X. (2014). First report of Maize chlorotic mottle virus infecting sugarcane (Saccharum officinarum). Plant Disease, 98, 572–572.10.1094/PDIS-07-13-0727-PDN
- Wen, W. G., Zhang, J. C., Cui, J. X., & Zhang, Y. (2011). Detection of Maize chlorotic mottle virus by real-time fluorescent RT-PCR. Plant Quarantine [in Chinese], 25, 39–42.
- Wu, J. X., Wang, Q., Liu, H., Qian, Y. J., Xie, Y., & Zhou, X. P. (2013). Monoclonal antibody-based serological methods for Maize chlorotic mottle virus detection in China. Journal of Zhejiang University Science B, 14, 555–562.10.1631/jzus.B1200275
- Yue, Z. Q., Teng, Y., Liang, C. Z., Xie, X. Y., Xu, B., Zhu, L. H., … Jiang, Y. L. (2008). Development of a sensitive and quantitative assay for spring viremia of carp virus based on real-time RT-PCR. Journal of Virological Methods, 152, 43–48.10.1016/j.jviromet.2008.05.031
- Zeng, C., Huang, X., Xu, J. M., Li, G. F., Ma, J., Ji, H. F. … Chen, H. J. (2013). Rapid and sensitive detection of Maize chlorotic mottle virus using surface plasmon resonance-based biosensor. Analytical Biochemistry, 440, 18–22.10.1016/j.ab.2013.04.026
- Zhang, Y., Hung, T., Song, J. D., & He, J. S. (2013). Electron microscopy: Essentials for viral structure, morphogenesis and rapid diagnosis. Science China Life Sciences, 56, 421–430.10.1007/s11427-013-4476-2
- Zhang, Y. J., Zhao, W. J., Li, M. F., Chen, H. J., Zhu, S. F., & Fan, Z. F. (2011). Real-time TaqMan RT-PCR for detection of maize chlorotic mottle virus in maize seeds. Journal of Virological Methods, 171, 292–294.10.1016/j.jviromet.2010.11.002