Abstract
African indigenous cattle breeds have been reared within the continent for millennia. Due to the harsh tropical environmental conditions (e.g. sever disease and parasite prevalence, high temperature, feed and water scarcity) where they evolved, they have developed various levels of tropical environment adaptation attributes. In order to explore the genomic signatures of tropical environment adaptation in African cattle, we compared the whole genomes of East African Indicus cattle breeds with European and Asian Taurine cattle breeds using XP-EHH and XP-CLR population statistical methods. Several genes involved in various biological processes and pathways related to domestication and behavior (dopaminergic and glutamatergic synapse), feeding and metabolism (gastric acid secretion, metabolic pathways), thermotolerance (sphingolipid and Wnt signaling), immune system response (T cell receptor signaling), and growth and reproduction (osteoblast differentiation, fibroblast migration) were identified from our genome analysis. Genes associated with tick and parasite resistance traits such as keratin genes, collagen genes, calcium signaling, and tumor necrosis factor proteins were also identified. The genes and pathways identified in this study improve our understanding of the biological mechanisms of tropical environment adaptation of African Indicus cattle breeds, which may allow us to use them for genomic selection programs. This result presents a basis for further study and may help to develop vaccines for tick and gastrointestinal parasite challenge.
PUBLIC INTEREST STATEMENT
Cattle are an important livestock species that provide food (milk, meat, blood), fiber (hide), power (as a draft animal), and security for the smallholder poor farm households in developing countries including Africa. African cattle are known for their adaptation to tropical environmental conditions like thermotolerance, resistance to tropical diseases and have the ability to thrive under poor quality feed and water conditions. This is because, they are evolved under tropical environmental conditions of high year round temperature, poor quality and availability of feed and water, high prevalence of infectious and vector-borne diseases. Characterizing the genome of adaptive indigenous cattle populations and identifying genomic regions contributing to tropical environment adaptation is an important step in developing a breeding strategy for increased meat and milk production. Accordingly, in this study, we analyzed the genome of African Indicus cattle breeds and identified genomic regions that are associated with tropical environmental adaptation traits including light and shiny coat color, better sweating ability, higher feeding efficiency, better reproductive performances, and superior disease and parasite resistance.
1. Introduction
Sub-Saharan Africa is endowed with diverse (>145 cattle breeds) and a huge number of cattle populations (Rege, Citation1999). The genetic makeup of African cattle breeds is comprised primarily of genetic information originating from two subspecies: the humpless B. taurus (taurine) and the humped B. indicus (indicine). The contribution of the North African subspecies of wild cattle or aurochs B. primigenius to the genome of some African cattle populations is a possibility as well (Decker et al., Citation2014). Taurine cattle, known to have been domesticated in the near east and introduced to Africa, are the earliest African cattle which are, nowadays, inhabiting mainly the humid and sub-humid areas in West and Central Africa. Indicine cattle, domesticated in the Indus valley, were introduced to Africa later most likely after 700 AD, during Arabic migrations into north and east Africa (Ajmone Marsan, Garcia, & Lenstra, Citation2010; Hanotte et al., Citation2002; Rege, Citation1999). These days African cattle breeds are the result of human and natural selection forces, and introgression between the subspecies has shaped their genetic diversity resulting in breeds of diverse morphological and physiological characteristics, as well as adaptation to different agro-ecological and sociocultural conditions (Bahbahani, Afana, & Wragg, Citation2018; Bahbahani et al., Citation2015, Citation2018; Hanotte, Dessie, & Kemp, Citation2010; Mwai, Hanotte, Kwon, & Cho, Citation2015; Rege, Citation1999).
Environmental adaptation attributes including disease and parasite resistance as well as heat tolerance are important traits for cattle survival and production in tropical and subtropical regions (Neto, Jonsson, Michael, & Barendse, Citation2011; Shyma, Gupta, & Singh, Citation2015). High prevalence of disease and parasites exert a significant effect on cattle production in Africa causing death and production losses (Mwai et al., Citation2015). External parasites such as ticks, which feed on blood and transmit various types of diseases impact livestock production and productivity. The estimated economic cost of tick infestation control is high (Mapholi et al., Citation2014; Neto et al., Citation2011). Similarly, high environmental temperature compromises animal production and productivity through its effect on growth, reproduction, and feed intake (Hansen, Citation2004). Moreover, the stringency of available environmental resources in tropical regions make the introduction of productive European breeds difficult (Wang et al., Citation2007).
Humped zebu cattle are Indicus cattle which are abundantly distributed throughout East Africa and the drier parts of West Africa. These cattle breeds are easily identified by their characteristic hump and pendulous dewlap and are thought to have descended from the secondary cattle introduction that occurred in the arid regions of Africa. Zebu cattle are known for their superior adaptation to harsh environments, including resistance to various types of diseases and parasites as well as thermotolerance (Bahbahani et al., Citation2015, Citation2018; Chan, Nagaraj, & Reverter, Citation2010; Kongsuwan et al., Citation2010; Rege, Citation1999).
Characterizing the genome of indigenous cattle populations and identifying genomic regions contributing to tropical environment adaptation is an important step in developing a breeding strategy for increased meat and milk production (Bahbahani et al., Citation2015, Citation2018; Chan et al., Citation2010). Exploring signatures of positive selection has been used to identify genomic regions affected by natural and artificial selection that are associated with the phenotypes of specific populations. Previously, several authors have reported the positive selection of genes associated with tick resistance and other environmental adaptation traits in tropical cattle breeds. For example, Chan et al. reported genes directly and/or indirectly associated with tropical adaptation attributes such as keratins, heat shock proteins, and heat tolerance genes (Chan et al., Citation2010). By comparing the differential expression of genes in resistant and susceptible cattle, several candidate genes responsible for tick resistance were identified (Kongsuwan et al., Citation2010; Piper et al., Citation2008; Wang et al., Citation2007). The major histocompatibility complex genes have been the center of focus for genetic variation of disease resistance studies in cattle (Martinez et al., Citation2006; Neto et al., Citation2011). Gautier et al. reported genes involved in the immune system, nervous system, and skin and hair properties in West African cattle breeds (Gautier et al., Citation2009). Several genes involved in various biological pathways such as immunity, reproduction, development and heat tolerance have also been reported for East African Shorthorn Zebu cattle (Bahbahani et al., Citation2015, Citation2017) and Ethiopian Sheko cattle (Bahbahani et al., Citation2018). However, studies on East African cattle breeds based on whole genome next-generation sequencing (NGS) data are very scarce.
Several studies reported the positive selection signature of genes in various cattle breeds using different methods (Bahbahani et al., Citation2018, Citation2015, Citation2017; Gautier et al., Citation2009; Lee et al., Citation2014; Noyes et al., Citation2011). Here, we used a cross-population extended haplotype homozygosity (XP-EHH) and cross-population composite likelihood ratio (XP-CLR) statistical methods. The XP-EHH statistic assesses haplotype differences between two populations. It is designed to detect alleles that have increased in frequency to the point of fixation or near fixation in one of the populations being compared (Pickrell et al., Citation2009; Sabeti et al., Citation2007). XP-CLR is a likelihood method for detecting selective sweeps that involve jointly modeling the multilocus allele frequency differentiation between two populations (Chen, Patterson, & Reich, Citation2010). Using these two statistics, we compared the whole genomes of three East African Indicine cattle breeds with four (Three European and one Asian) Taurine cattle breeds in order to identify positively selected genomic regions in African cattle breeds that are associated with tropical environmental adaptation traits.
2. Materials and methods
2.1. Data description and preparation
This study is based on previously published data; detailed information about sampling and re-sequencing of DNA samples can be found within the manuscript (Kim et al., Citation2017). DNA samples extracted from whole blood samples of three East African Indicus cattle breeds (9 Boran, 9 Ogaden, and 10 Kenana) were individually sequenced using the Illumina HiSeq 2000 platform. Additionally, previously published genomic data of three European (10 Holstein, 10 Angus, and 10 Jersey) and a single Asian (11 Hanwoo) Taurine cattle breeds was used. Together, a per-base sequence quality check was performed using the FastQC software (Andrews, Citation2010). Bowtie2 (Langmead & Salzberg, Citation2012) was used to map pair-end sequence reads to the reference bovine genome (UMD 3.1). There was an overall alignment rate of 98.5% and average read depth of 10.8x. Picard tools (http://picard.sourceforge.net) to filter potential PCR duplicates, and SAMtools was used to create index files for reference and bam files (Li et al., Citation2009). Genome analysis toolkit 1.4 (GATK) performed local realignment of reads (McKenna et al., Citation2010). The “UnifiedGenotyper” and “SelectVariants” arguments of GATK were used to call candidate SNPs. In order to filter variants and avoid possible false positives, the “VariantFiltration” argument of the same software was adopted using the following options: 1) SNPs with a phred-scaled quality score of less than 30 were filtered; 2) SNPs with MQ0 (mapping quality zero; total count across all samples of mapping quality zero reads) >4 and quality depth (unfiltered depth of non-reference samples; low scores are indicative of false positives and artifacts) <5 were filtered; and 3) SNPs with FS (Phred-scaled p-value using Fisher’s exact test) > 200 were filtered since FS represents variation on either the forward or the reverse strand, which is indicative of false positive calls. BEAGLE (Browning & Browning, Citation2007) was used to infer the haplotype phase and impute missing alleles for the entire set of cattle populations simultaneously. After performing all the filtering processes, a total of ~37 million SNPs were retained and used for further analysis in this study. The sequences used in this study are available from GenBank with the Bioproject accession number of African cattle breeds (PRJNA312138—SRX1858079, SRX1858078, SRX1858077, SRX1858076, SRX1858075, SRX1858074, SRX1858073, SRX1858072, SRX1858071, SRX1802254, SRX1802253, SRX1802251, SRX1775710, SRX1775700, SRX1766099, SRX1766098, SRX1766097, SRX1766096, SRX1765344, SRX1765338, SRX1762810, SRX1762809, SRX1752210, SRX1751992, SRX1640149, SRX1631639, SRX1625858, SRX1765311), Holstein (PRJNA210521—SRX322362, SRX322361, SRX322360, SRX322359, SRX322358, SRX322357, SRX322356, SRX322355, SRX322354, SRX322353), Jersey (PRJNA318089—SRX1756427, SRX1756425, SRX1756315, SRX1756311, SRX1756308, SRX1756302, SRX1756292, SRX1756266, SRX1756143, SRX1756073), Angus (PRJNA318087—SRX1762572, SRX1762571, SRX1762569, SRX1762564, SRX1762377, SRX1762017, SRX1762016, SRX1758657, SRX1757535, SRX1756016), and Hanwoo (PRJNA210523—SRX322347, SRX322352, SRX322351, SRX322350, SRX322349, SRX322348, SRX322346, SRX322345, SRX322344, SRX318521, SRX318497).
2.2. Phylogenetic tree and population structure
In order to infer the evolutionary relationship between the breeds considered, we constructed a phylogenetic tree using genome-wide autosomal SNPs from 69 individual sequences. We used the SNPhylo pipeline (Lee, Guo, Wang, Kim, & Paterson, Citation2014) to construct the phylogenetic tree with a sub sample of 17,243 SNPs and options of Minor allele frequency (MAF) >0.05 and number of bootstrap samples of 1000. FigTree (http://tree.bio.ed.ac.uk/software/figtree/) was used to visualize the tree. Additionally, we used STRUCTURE software which implements Bayesian algorithms in order to detect the true number of clusters, K (the number of ancestral populations), to identify groups of individuals corresponding to the uppermost hierarchical levels (Evanno, Regnaut, & Goudet, Citation2005). A total number of 16,181 loci were used for the structure analysis, using the following options of Length of Burnin Period of 2000, Number of MCMC Reps after Burnin of 100,000 and MAF of 0.05. We used VCFtools (Price et al., Citation2006) and PLINK (Purcell et al., Citation2007) with thin option (0.000675) to prepare input data used by STRUCTURE.
2.3. Detection of signature of positive selection
In order to detect the positive selective sweep regions in East African Indicus cattle, the whole genomes of three East African Indicus cattle breeds—Boran (Kenya), Ogaden (Ethiopia), and Kenana (Sudan)—grouped together as a test population were compared with four (three European and one Asian) Taurine cattle breeds—Holstein, Angus, Jersey and Hanwoo—grouped together as a reference population. East African Indicus cattle breeds are the result of the second introduction of Zebu cattle to Africa and have been bred for millennia under the harsh African conditions. Therefore, comparing the genomes of these breeds with European and Asian breeds and detecting recent positive selection signatures might give an insight into the selective forces shaping the genomes that help the African cattle adapting to the harsh environmental conditions.
We used the XP-EHH statistical method to assess haplotype differences between the test and reference populations (Sabeti et al., Citation2007). XP-EHH compares haplotype lengths between populations to control for local variation in recombination rates. It detects alleles that have increased in frequency to the point of fixation or near fixation in one of the populations (Pickrell et al., Citation2009). XP-EHH is directional, meaning that the direction of selection is determined by the sign of the values; positive values indicate selective sweep in the test populations, whereas a negative value indicates selection in the reference populations. We followed previously published procedures (Kim, Cho, Caetano-Anolles, Kim, & Ryu, Citation2015; Pickrell et al., Citation2009). XP-EHH values were calculated using the XP-EHH software, which can be accessed through the following link: (http://hgdp.uchicago.edu/Software/). In order to compare the genomic regions across populations, the genome was split into non-overlapping segments of 50 kb and the maximum XP-EHH value was computed for each segment. In order to define the empirical P-value, segments were grouped into three clusters according to the number of SNPs. In each cluster, the regions with P-values less than 0.01 (1%) were considered as strong signals of selection in the African cattle populations.
Additionally, we performed an XP-CLR statistical test in order to identify potential regions differentially selected between the two populations compared (Chen et al., Citation2010). XP-CLR is a likelihood method for detecting selective sweep regions based on allele frequency differentiation. The script available at (http://genepath.med.harvard.edu/reich) was used to calculate XP-CLR scores. Non-overlapping sliding windows of 50 kb and a maximum number of 600 SNPs within each window were used. The correlation level from which the SNPs contribution to XP-CLR result was down-weighted to 0.95. The top 1% (0.01) of the empirical distributions were designated as candidate sweeps and genes that span the window regions were defined as candidate genes (Kim et al., Citation2015). Significant genomic regions identified from the XP-EHH and XP-CLR tests were annotated based on UMD 3.1.
2.4. Characterization of candidate genes under selection
The genes identified from XP-EHH and XP-CLR statistics were submitted to the Database for Annotation, Visualization, and Integrated Discovery (DAVID) gene ontology and annotation tool for gene enrichment analysis (Huang, Sherman, & Lempicki, Citation2009). Gene Ontology (GO) Biological Process (BP) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of the DAVID tool were used to cluster the genes of similar biological functions and pathways. All genes identified from XP-EHH and XP-CLR were submitted together.
We used SNPEff, a genetic variants (amino acid changes) annotation and effect prediction tool, for the candidate genes (Cingolani et al., Citation2012). Missense variants of candidate genes were extracted and carried on for the logistic and association analysis. By employing the Chi-squared test (plink—file mydata—assoc option) and logistic model (plink—file mydata—logistic option) employed in PLINK V1.07 software, significant SNPs that are specific for African Indicus cattle were identified and highly significant variants are reported.
The gene names and descriptions used in this manuscript are based on genecards (http://www.genecards.org/). The Manhattan plots of the—log10 transformed XP-EHH values and XP-CLR scores were drawn using R software.
3. Result and discussion
3.1. Phylogenetic tree and population structure
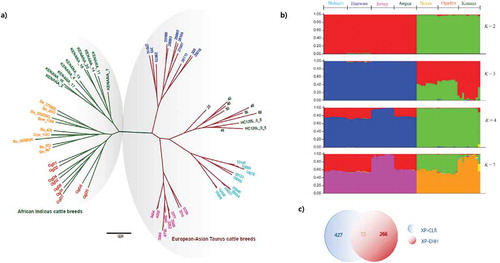
In order to assess the historical relationship among the cattle breeds considered, we constructed a non-rooted phylogenetic tree of 69 individual cattle using autosomal SNPs. As expected, the phylogenetic tree revealed that African Indicus cattle breeds are clustered together separately from European and Asian Taurine cattle breeds (Figure ). Additionally, we performed population structure (Evanno et al., Citation2005) at different population assumptions, K (Figure ; Additional file 2: Figure S1). When K is 2, consistent with the phylogenetic tree, African Indicine and European and Asian Taurine cattle breeds showed clear differences, but at K 3, African Indicus breeds showed an admixture of each other. At K ≥ 4 population assumptions, except Jersey cattle, European and Asian Taurine cattle breeds showed some admixture level. In general, our result is consistent with previous reports which indicated that Indicine cattle are divergent from Taurine cattle breeds (Decker et al., Citation2014).
Figure 1. Population structure and relationship of cattle breeds considered. (a) Phylogenetic tree (Green Branch, Green Node—Kenana; Green Branch, Orange Node—Boran; Green Branch, Red Node—Ogaden; Red Branch, Blue Node—Hanwoo; Red Branch, Green Node—Angus; Red Branch, Cyan Node—Holstein; Red Branch, Rose Node, (b) Population structure at 2, 3, 4 and 7 population assumptions, (c) Number of genes identified from XP-CLR and XP-EHH statistics.
3.2. Signature of positive selection in African Indicus cattle populations
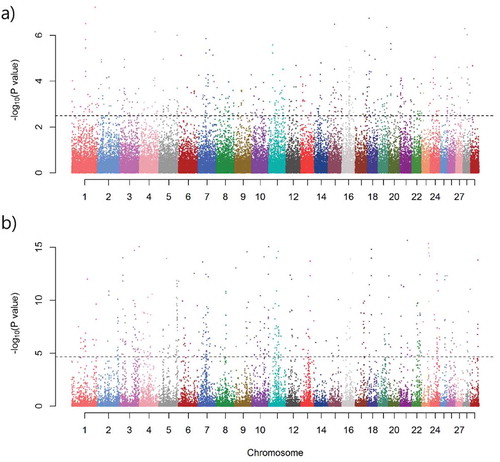
In order to detect positive selection sweep regions in East African Indicus cattle populations, we used XP-EHH (Pickrell et al., Citation2009; Sabeti et al., Citation2007) and XP-CLR (Chen et al., Citation2010) population statistical methods. We compared the genomes of three East African Indicus cattle breeds (grouped together as a test population) with the genomes of four (three European and one Asian) Taurine cattle breeds (grouped together as a reference population). The Manhattan plot of the -log10 transformed XP-CLR and XP-EHH P-values is presented in Figure . From the signature of selection analysis, 257 (XP-EHH statistics) and 345 (XP-CLR statistics) outlier (1%) selective sweep genomic regions were identified under selection. Annotation of these outlier regions resulted in a total of 765 (XP-EHH = 338; and XP-CLR = 499) genes (Figure ; Additional file 1: Table S1-S2).
Figure 2. Manhattan plot of the—log10 transformed XP-EHH (a), and XP-CLR (b) p-values. The y-axis shows the—log10 (P-value) of XP-EHH and XP-CLR p-value, and x-axis shows chromosomal positions. The horizontal dotted lines represent the 1% outlier regions in both of the statistical methods.
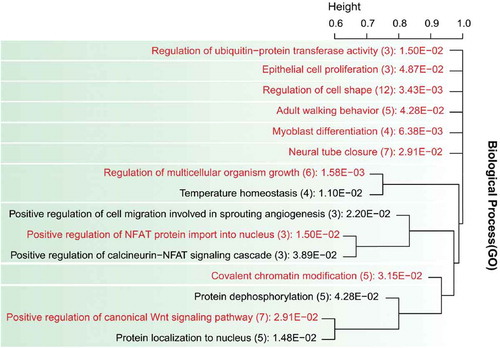
Next, we performed DAVID gene enrichment analysis to identify enriched GO BP terms and KEGG pathways (Huang et al., Citation2009) using all the genes (765) identified from both statistics, and 15 significant (p < 0.05) BP terms and 19 KEGG pathways were found enriched. We clustered significantly enriched BP terms based on genes involved in the terms (Figure ). These BP terms are involved in various biological functions including immune response (NFAT signaling cascade), growth and development (cell proliferation, cell shape, myoblast differentiation, neural tube closure, canonical Wnt signaling pathway), and behavior (walking behavior).
Figure 3. Functional clustering of Gene Ontology Biological Process terms. The BP terms are associated with genes located in outlier loci (regions) detected by XP-CLR and XP-EHH statistics in East African Indicus cattle breeds. The gene list of each GO term in the cluster was compared to calculate the distance between the GO terms. For a distance of >0.8–0.9, GO terms were re-clustered. The representative GO terms in a group were manually selected, and are shown in red font. The numbers of genes in the GO terms are in brackets, along with their corresponding p-value.
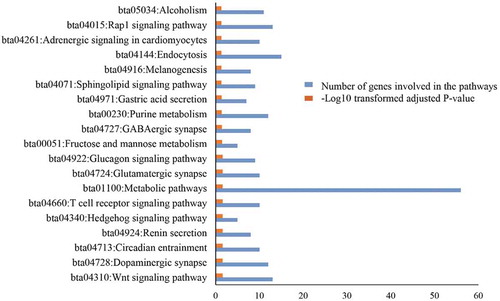
Enriched KEGG pathways are presented in Figure (see Additional file 2: Table S3 for genes in the pathways). Dopaminergic synapse was among the KEGG pathways enriched. Dopamine is an organic chemical of the catecholamine and phenethylamine families that mediates a wide range of brain functions. It is involved in the domestication process of animals (Nikulina, Citation1990). Glutamatergic synapse pathway plays an important role in the behavioral adaptation of stress and fear responses (Kamprath et al., Citation2009). These pathways might also be related to domestication, which effects tameness and fear response in domestic animals (Mirkena et al., Citation2010; Zeder, Citation2012). Dopamine and glutamate modulate foraging and feeding-related behaviors of animals (Hills, Citation2006). The dopaminergic system aids free-ranging animals in identifying grass/forage species that are nutritious and non-poisonous (Berthoud, Citation2007; Jensen, Citation2002). The pastoral mode of livestock production in Africa might contribute for the positive selection of the gene regions involved in these pathways. African pastoralists move from place to place with their cattle in search of feed and water, which forces cattle to adapt to new vegetation and feed types (Hanotte et al., Citation2002; Mwai et al., Citation2015).
Figure 4. KEGG pathways enriched from positively selected identified genes. The y-axis is the pathways and the numbers in the x axis are number of genes that are included in the KEGG pathway enriched and the log10 transformed adjusted p-values.
The KEGG pathways of gastric acid secretion, and fructose and mannose metabolism enriched are related to feeding digestion and metabolism. Gastric acid is vital for normal upper gastrointestinal functions, including protein digestion, and calcium and iron absorption, as well as providing some protection against bacterial infections. The physical and chemical nature of livestock feed affects gastric function to which coarse textured feeds result in higher gastric acid secretion (Low, Citation1990). The poor nutritional quality and seasonal availability of livestock feed in tropical Africa can act as a selective pressure which contributes to the adaptation and efficient utilization of low-quality feed by indigenous African cattle breeds. Lower metabolic requirements or reducing metabolism and higher digestive efficiency, and ability to utilize poor quality feed are adaptations of tropical cattle to feed scarcity (Mirkena et al., Citation2010). The higher efficiency of Indicus cattle to ferment nitrogen-deficient poor-quality feed and capture energy has been reviewed (Hegarty, Citation2004; Warwick & Cobb, Citation1975).
The mammalian heat shock response involves the sphingolipid signaling pathway, which activates sphingomyelin hydrolysis and/or the de novo biosynthesis of sphingolipids (Jenkins, Citation2003). The sphingolipid signaling pathway has two main metabolites: the ceramide and sphingosine-1-phosphate, which are important for heat stress response. Ceramides signals cells to undergo apoptosis during a severe heat stress (Jenkins, Citation2003). Sphingosine-1-phosphate has been found to protect oocytes from physiologically relevant heat shock and affect oocyte maturation (Roth & Hansen, Citation2004). Wnt signaling is also required for sweat gland development and sweating (Cui et al., Citation2014). Evaporative cooling through the involvement of sweat glands and other skin components is an important mechanism of heat tolerance (Jian, Duangjinda, Vajrabukka, & Katawatin, Citation2014). The genes in these pathways (PLCB1, MAPK12, and SGPL1) have been found to be involved in different aspects of thermotolerance. PLCB1 is associated with heat tolerance in catfish (Jin et al., Citation2017) and has been taken as an adaptation to hot arid environments in sheep and goats (Kim et al., Citation2016). Mitogen-activated protein kinases (MAPKs) elicit cellular response favoring survival or apoptosis (Sugimoto et al., Citation2012). Renin secretion, enriched in the KEGG pathways, is associated with adaptation to heat stress (Ali et al., Citation2012). It regulates the body’s water balance through the secretion of aldosterone and animals adapted to hot arid environments display lower overall water intake and turnover (Mirkena et al., Citation2010; Warwick & Cobb, Citation1975). Additionally, heat shock protein families (DNAJC11, DNAJC8), heat shock factor proteins (HSF5) and genes associated with heat stress response (PPP2R5E) identified in this study might contribute to the superior heat tolerance ability of African Indicus cattle breeds (Bahbahani et al., Citation2018, Citation2015, Citation2018, Citation2017; Wang, Dzama, Rees, & Muchadeyi, Citation2015). The BP term temperature homeostasis, a strategy by the animal to cope with climate variability, was also enriched.
Renin is found in high concentrations in reproductive tissues indicating its role in reproduction function. It affects ovulation, angiogenesis, and steroidogenesis (Yoshimura, Citation1997). Hedgehog signaling is an important pathway for proper development of the reproductive system (Franco & Yao, Citation2012). It is implicated in male germ cell development and differentiation in chicken (Chen et al., Citation2017). In addition to pathways, genes involved in reproduction function such as IGF-1, ESR2, and FGFR2 were identified (Table ). IGF-1 affects follicular development and oocyte maturation and also enhances ovarian angiogestin II production (Yoshimura, Citation1997). ESR2 is a protein-coding gene that controls many cellular processes including growth, differentiation, and function of the reproductive system. A polymorphism in ESR2 gene has been shown to be associated with sperm quality and boar fertility traits (Gunawan et al., Citation2012). FGFR2 is among the mitogenic signaling molecules with an essential role in the regulation of embryonic development. It is involved in postnatal development and function of the uterus (Filant, DeMayo, Pru, Lydon, & Spencer, Citation2013). The significant effect of high temperature on reproductive function of livestock species has been previously observed (Hansen, Citation2009). The pathways and genes identified here might contribute to the better reproductive efficiency of African Indicus cattle breeds to produce and reproduce in the tropical harsh environmental conditions of the continent (Hansen, Citation2009; Makina et al., Citation2015).
Table 1. Candidate genes identified as positively selected in African cattle breeds affecting different traits in relation to tropical environment adaptation
The T cell receptor signaling pathway, an important pathway for immune system development, was identified in this study. T cell receptors play a key role in the functioning of T cells and formation of an immunological synapse between T cells and the antigen-presenting cells. They initiate activation of T cells which in turn causes clonal expansion, differentiation, cytotoxic killing, or induction of programmed cell death (Bromley et al., Citation2001). The positive selection of genes involved in gene ontologies and pathways related to immune response have been previously reported for African cattle (Makina et al., Citation2015; Wang et al., Citation2015). Under tropical environments, indigenous African cattle breeds tend to better resist disease and parasite challenge as compared to European cattle breeds (Wang et al., Citation2015). The high and diverse disease and parasite prevalence in tropical Africa (Mirkena et al., Citation2010) where these breeds have evolved might be the selective pressure for the genes involved in pathways and BP terms related to immune responses.
Additionally, genes that contribute to the superior tick and parasite resistance mechanisms in African Indicus cattle were identified (Table ). Host tick resistance is defined as the ability of the host to reduce damage caused by the parasite through diverse immune and non-immune structural components (Neto et al., Citation2011). Immune response genes identified in relation to tick resistance include BoLA, TNFAIP8L3, and SLC25A48. BoLA, also known as the bovine major histocompatibility complex, has a major role in antigen processing and presentation. It plays a vital role in host resistance to an ecto- and endoparasite infestation (Martinez et al., Citation2006; Shyma et al., Citation2015). BoLA is highly polymorphic; several scholars reported the association of different alleles to tick resistance (Martinez et al., Citation2006; Neto et al., Citation2011). TNFAIP8L3 and SLC25A48 genes are involved in antigen recognition and have been previously identified to be under selection in African cattle breeds in relation to tick resistance (Makina et al., Citation2015).
The physical structure of the epidermal layers of cattle skin represents the first line of defense against ectoparasite invasion (Gautier et al., Citation2009; Kongsuwan et al., Citation2010). In relation to this, keratin and keratin associated genes (KRT33A, KRTAP27-1, and KRTAP9-1) were identified under selection. Keratin genes are heteropolymeric structural proteins that form the structural framework of the skin and hair cells contributing to tick resistance via their action as a barrier to the external environment (Nakamura, Kanemarum, & Fukami, Citation2013). KRT33A gene was previously found overrepresented in highly tick resistant cattle (Kongsuwan et al., Citation2010). Keratinocytes in the epidermis also have an immune response function- they secrete cytokines that initiate local inflammatory responses (Nakamura et al., Citation2013). Tick resistance in African cattle has been understood to be due to superior skin immunity (Marufu, Qokweni, Chimonyo, & Dzama, Citation2011). The positive selection of keratin-related genes has been previously reported in African cattle (Bahbahani et al., Citation2015; Chan et al., Citation2010; Makina et al., Citation2015). Collagens (COL12A1, COL8A1) provide structural integrity to the skin (Wang et al., Citation2007). Structural integrity was found overrepresented from genes differentially expressed in tick resistant Brahman cattle (Kongsuwan et al., Citation2008). Extracellular matrix proteins (PRG3, VWA2, and ATRN) form a protective barrier producing an unfriendly environment for tick attachment and feeding (Kongsuwan et al., Citation2008). PRG3 stimulates neutrophil superoxide production and histamine release from basophils. It is involved in histamine biosynthetic process and activates basophils that are important effectors of tick rejection and a major component of acquired resistance of the host (Falcone, Pritchard, & Gibbs, Citation2001; Wikel, Citation1996).
The contribution of light coat color to cattle tick resistance has been previously reported (Ibelli et al., Citation2012; Mapholi et al., Citation2014; Marufu et al., Citation2011). This might be due to the fact that on light colored animals ticks can be easily seen by predator birds and picked up easily (Ibelli et al., Citation2012; Mapholi et al., Citation2014; Marufu et al., Citation2011). Melanogenesis, defined as the production of melanin pigments, plays a role in the pigmentation of skin and hair (Cieslak, Reissmann, Hofreiter, & Ludwig, Citation2011). Several genes (SLC45A2, MLPH, RAB17, RAB37, RAB42, RAB7A, and ATRN), which are involved at different stages of melanocyte biology (Guibert, Girardot, Leveziel, Julien, & Oulmouden, Citation2004), were detected in this study. Mutations in these genes cause dilution of phenotypes (Cieslak et al., Citation2011), which might result in light coat color of African Indicine cattle breeds (Marufu et al., Citation2011). Mutations in MLPH gene have been found to cause dilution of coat color (Philipp et al., Citation2005). In addition to color, cattle hairs are coated with an emulsion of sweat and sebum, which prevents tick attachment (Kongsuwan et al., Citation2010). Searching for non-synonymous mutations, we identified five (three known—rs136185632, rs209852661, rs209010309 and two novel—3:117616155, 3:117632216), and two known (rs134604394, rs378039235) significant missense variants on MLPH and SLC45A2 gene regions, respectively (Table ). MC5R controls the secretion of sebum (Zhang, Li, Anthonavage, & Eisinger, Citation2006), a complex mixture of lipid that contributes to skin integrity and inflammatory processes, transport antioxidants to the skin surface, and has innate antimicrobial activity (Zhang et al., Citation2011). In this gene region, one novel (24:43984677) and one known (rs134437472) significant missense variants were identified (Table ). The smoother coat and shorter hair of indigenous African cattle breeds have been reported to contribute to their superior tick resistance (Marufu et al., Citation2011).
Table 2. Significant missense variants identified in candidate gene regions
Genes involved in calcium signaling (ATP2A2, TGM3, and TGM1) have also been found to play an important role in host response to tick challenge (Bagnall, Gough, Cadogan, Burns, & Kongsuwan, Citation2009; Kongsuwan et al., Citation2010, Citation2008). The ATP2A2 gene, which encodes Ca2+dependent ATPase, is reported to be highly expressed in highly tick resistant cattle (Bagnall et al., Citation2009). Transglutaminases (TGM3, TGM1) are Ca2+dependent enzymes expressed in terminally differentiating keratinocytes that are involved in the apoptosis and cornification of the epidermis. TGMs are required for crosslinking the main component of the epidermal cornified envelope in the epidermis (Candi, Schmidt, & Melino, Citation2005) and provide a defense function against ectoparasites (Kongsuwan et al., Citation2010). In TGM1 and TGM3 gene regions, we identified two novel (10:20736590, 10:20739046) and three known (rs41695720, rs136283113, rs211468449) significant missense variants, respectively (Table ).
Gastrointestinal (GI) parasites are economically significant livestock health problems causing production losses (Benavides, Sonstegard, & Van Tassell, Citation2016; McManus, Do Prado Paim, de Melo, Brasil, & Paiva, Citation2014). Gastric acid secretion, one of the identified enriched KEGG pathways, has a role in protecting the host against several parasitic diseases in humans (Martinsen, Bergh, & Waldum, Citation2005). Genes involved in GI parasite resistance identified in this study include TNFAIP3, TNFAIP8L3, and DMBT1. TNF proteins encode a multifunctional pro-inflammatory cytokine related to nematode resistance (Araujo et al., Citation2009). DMBT1 is a protein-coding gene that plays a crucial role in mucosal defense, cellular immune defense, and epithelial differentiation. It has been found highly expressed in the mesenteric lymph node of intestinal nematode resistant animals (Araujo et al., Citation2009). Through observation of the genetic variation relating to gastrointestinal parasite resistance between and within breeds, it is evident (McManus et al., Citation2014) that Indicus cattle have evolved for superior parasite resistance as compared to Taurine cattle and their crosses (Frisch, O’neill, & Kelly, Citation2000).
4. Limitation of this study
As false positive results are not uncommon in these kinds of studies, additional validation procedures are important. Additionally, although the statistical methods used are robust enough to identify recent signatures due to natural and artificial selection, the genes identified here might be due to the divergence between the two species which occurred millions of years ago.
5. Conclusion
Results of our analysis identified important putative genes and pathways that are positively selected in East African Indicus cattle in response to different selection pressures of the African tropical environment. The interaction of African cattle breeds with the high disease and parasite prevalence, high temperature, and seasonal feed and water scarcity prevailing in the region might be the main selective pressure underlying the selection signature of genes. Identification of these genes allows us to better understand the biological process and mechanisms of tropical environmental adaptation in African cattle and can be used in future genomic selection programs. The genes identified in relation to tick and parasite resistance can be used for the development of tick and parasite control methods such as designing selection programs and development of vaccines.
Additional file
Additional file 1: Table S1. List of genes identified as positively selected in African cattle from a genome-wide comparison of three East African Indicine cattle breeds and four (three European and one Asian) Taurine cattle breeds using XP-CLR statistics; Table S2. List of genes identified as positively selected in African cattle from a genome-wide comparison of three East African cattle breeds and four (three European and one Asian) Taurine cattle breeds using XP-EHH statistics (Common genes are indicated with red text).
Additional file 2: Table S3. Enriched KEGG Pathways overrepresented from DAVID Gene ontology analysis for genes identified from East African Indicus cattle breeds. Table S4. Gene name and description of candidate genes that contribute to tropical environment adaptation mechanisms of East African Indicine cattle (from genecards [provided by RefSeq, Apr 2014] (http://www.genecards.org/) and literature). Figure S1. Population structure at K = 3, 5 and 6 population assumptions in seven cattle breeds.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Supplemental Material
Download Zip (172.1 KB)Acknowledgements
This work was supported by Agenda (PJ01134905) of the National Institute of Animal Science, Rural Development Administration (RDA), Korea.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
Notes on contributors
Mengistie Taye
Mengistie Taye The first (corresponding) author is a PhD holder specialized in Agriculture Biotechnology. His PhD research was focusing on bioinformatics analysis of the genomes of animals for adaptive signature of natural and artificial selection in cattle. He has published his findings in different peer reviewed international journals. Analysis of signature of selection is a genotype to phenotype approach and helps to identify and understand whether the phenotypic traits manifested are due environmental acclimatization or genetic effects. Nowadays, there are a number of research projects working on the analysis of signature of selection for different adaptation and production traits in different species of animals. This manuscript, I believe, will add a lot to the knowledge base of population genetics and animal breeding in developing countries where adaptation traits are very important.
References
- Ajmone Marsan, P., Garcia, J. F., & Lenstra, J. A. (2010). On the origin of cattle: How aurochs became cattle and colonized the world. Evolutionary Anthropology, 19(4), 148–157. doi:10.1002/evan.20267
- Ali, M. A., Adem, A., Chandranath, I. S., Benedict, S., Pathan, J. Y., Nagelkerke, N., … Nicholls, G. M. (2012). Responses to dehydration in the one-humped camel and effects of blocking the renin-angiotensin system. PloS one, 7(5), e37299. doi:10.1371/journal.pone.0037299
- Andrews, S. (2010). FastQC: A quality control tool for high throughput sequence data. Reference Source.
- Araujo, R. N., Padilha, T., Zarlenga, D., Sonstegard, T., Connor, E. E., Van Tassel, C., … Gasbarre, L. C. (2009). Use of a candidate gene array to delineate gene expression patterns in cattle selected for resistance or susceptibility to intestinal nematodes. Veterinary Parasitology, 162(1), 106–115. doi:10.1016/j.vetpar.2008.12.017
- Bagnall, N., Gough, J., Cadogan, L., Burns, B., & Kongsuwan, K. (2009). Expression of intracellular calcium signalling genes in cattle skin during tick infestation. Parasite Immunology, 31(4), 177–187. doi:10.1111/j.1365-3024.2008.01092.x
- Bahbahani, H., Afana, A., & Wragg, D. (2018). Genomic signatures of adaptive introgression and environmental adaptation in the Sheko cattle of southwest Ethiopia. PloS one, 13(8), e0202479. doi:10.1371/journal.pone.0202479
- Bahbahani, H., Clifford, H., Wragg, D., Mbole-Kariuki, M. N., Van Tassell, C., Sonstegard, T., … Hanotte, O. (2015). Signatures of positive selection in East African Shorthorn Zebu: A genome-wide single nucleotide polymorphism analysis. Scientific Reports, 5. doi:10.1038/srep11729
- Bahbahani, H., Salim, B., Almathen, F., Al Enezi, F., Mwacharo, J. M., & Hanotte, O. (2018). Signatures of positive selection in African Butana and Kenana dairy zebu cattle. PloS one, 13(1), e0190446. doi:10.1371/journal.pone.0190446
- Bahbahani, H., Tijjani, A., Mukasa, C., Wragg, D., Almathen, F., Nash, O., … Woolhouse, M. (2017). Signatures of Selection for Environmental Adaptation and Zebu× Taurine Hybrid Fitness in East African Shorthorn Zebu. Frontiers in Genetics, 8, 68. doi:10.3389/fgene.2017.00068
- Benavides, M. V., Sonstegard, T. S., & Van Tassell, C. (2016). Genomic Regions Associated with Sheep Resistance to Gastrointestinal Nematodes. Trends in Parasitology, 32(6), 470–480. doi:10.1016/j.pt.2016.03.007
- Berthoud, H.-R. (2007). Interactions between the “cognitive” and “metabolic” brain in the control of food intake. Physiology & Behavior, 91(5), 486–498. doi:10.1016/j.physbeh.2006.12.016
- Bromley, S. K., Burack, W. R., Johnson, K. G., Somersalo, K., Sims, T. N., Sumen, C., … Dustin, M. L. (2001). The immunological synapse. Annual Review of Immunology, 19(1), 375–396. doi:10.1146/annurev.immunol.19.1.375
- Browning, S. R., & Browning, B. L. (2007). Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. The American Journal of Human Genetics, 81(5), 1084–1097. doi:10.1086/521987
- Candi, E., Schmidt, R., & Melino, G. (2005). The cornified envelope: A model of cell death in the skin. Nature Reviews: Molecular Cell Biology, 6(4), 328–340. doi:10.1038/nrm1619
- Chan, E. K., Nagaraj, S. H., & Reverter, A. (2010). The evolution of tropical adaptation: Comparing taurine and zebu cattle. Animal Genetics, 41(5), 467–477. doi:10.1111/j.1365-2052.2010.02053.x
- Chen, H., Patterson, N., & Reich, D. (2010). Population differentiation as a test for selective sweeps. Genome Research, 20(3), 393–402. doi:10.1101/gr.100545.109
- Chen, H, Zuo, Q, Wang, Y, Ahmed, M. F, Jin, K, Song, J, & Li, B. (2017). Regulation of hedgehog signaling in chicken embryonic stem cells differentiation into male germ cells (gallus). Journal Of Cellular Biochemistry, 118(6), 1379–1386. doi: 10.1002/jcb.25796
- Cieslak, M., Reissmann, M., Hofreiter, M., & Ludwig, A. (2011). Colours of domestication. Biological Reviews, 86(4), 885–899. doi:10.1111/j.1469-185X.2011.00177.x
- Cingolani, P., Platts, A., Wang, L. L., Coon, M., Nguyen, T., Wang, L., … Ruden, D. M. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly, 6(2), 80–92. doi:10.4161/fly.19695
- Cui, C.-Y., Yin, M., Sima, J., Childress, V., Michel, M., Piao, Y., & Schlessinger, D. (2014). Involvement of Wnt, Eda and Shh at defined stages of sweat gland development. Development (Cambridge, England), 141(19), 3752–3760. doi:10.1242/dev.109231
- Decker, J. E., McKay, S. D., Rolf, M. M., Kim, J., Alcalá, A. M., Sonstegard, T. S., … Praharani, L. (2014). Worldwide patterns of ancestry, divergence, and admixture in domesticated cattle. PLoS Genetics, 10(3), e1004254. doi:10.1371/journal.pgen.1004541
- Evanno, G., Regnaut, S., & Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Molecular Ecology, 14(8), 2611–2620. doi:10.1111/j.1365-294X.2005.02553.x
- Falcone, F. H., Pritchard, D. I., & Gibbs, B. F. (2001). Do basophils play a role in immunity against parasites? Trends in Parasitology, 17(3), 126–129.
- Filant, J., DeMayo, F. J., Pru, J. K., Lydon, J. P., & Spencer, T. E. (2013). Fibroblast growth factor receptor two (FGFR2) regulates uterine epithelial integrity and fertility in mice. Biology of Reproduction, 90(1), Article 7, 1–11.
- Franco, H. L., & Yao, H. H.-C. (2012). Sex and hedgehog: Roles of genes in the hedgehog signaling pathway in mammalian sexual differentiation. Chromosome Research, 20(1), 247–258. doi:10.1007/s10577-011-9254-z
- Frisch, J., O’neill, C., & Kelly, M. (2000). Using genetics to control cattle parasites—The Rockhampton experience. International Journal for Parasitology, 30(3), 253–264.
- Gautier, M., Flori, L., Riebler, A., Jaffrézic, F., Laloé, D., Gut, I., … Foulley, J.-L. (2009). A whole genome Bayesian scan for adaptive genetic divergence in West African cattle. BMC Genomics, 10(1), 1. doi:10.1186/1471-2164-10-550
- Guibert, S., Girardot, M., Leveziel, H., Julien, R., & Oulmouden, A. (2004). Pheomelanin coat colour dilution in French cattle breeds is not correlated with the TYR, TYRP1 and DCT transcription levels. Pigment Cell Research, 17(4), 337–345. doi:10.1111/j.1600-0749.2004.00152.x
- Gunawan, A., Cinar, M., Uddin, M., Kaewmala, K., Tesfaye, D., Phatsara, C., … Schellander, K. (2012). Investigation on association and expression of ESR2 as a candidate gene for boar sperm quality and fertility. Reproduction in Domestic Animals, 47(5), 782–790. doi:10.1111/j.1439-0531.2011.01968.x
- Hanotte, O., Bradley, D. G., Ochieng, J. W., Verjee, Y., Hill, E. W., & Rege, J. E. O. (2002). African pastoralism: Genetic imprints of origins and migrations. Science (New York, N.Y.), 296(5566), 336–339. doi:10.1126/science.1069878
- Hanotte, O., Dessie, T., & Kemp, S. (2010). Time to tap Africa’s livestock genomes. Science (New York, N.Y.), 328(5986), 1640–1641. doi:10.1126/science.1186254
- Hansen, P. (2004). Physiological and cellular adaptations of zebu cattle to thermal stress. Animal Reproduction Science, 82, 349–360. doi:10.1016/j.anireprosci.2004.04.011
- Hansen, P. (2009). Effects of heat stress on mammalian reproduction. Philosophical Transactions of the Royal Society of London, Series B: Biological Sciences, 364(1534), 3341–3350. doi:10.1098/rstb.2009.0131
- Hegarty, R. (2004). Genotype differences and their impact on digestive tract function of ruminants: A review. Animal Production Science, 44(5), 459–467. doi:10.1071/EA02148
- Hills, T. T. (2006). Animal foraging and the evolution of goal‐directed cognition. Cognitive Science, 30(1), 3–41. doi:10.1207/s15516709cog0000_50
- Huang, D. W., Sherman, B. T., & Lempicki, R. A. (2009). Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research, 37(1), 1–13. doi:10.1093/nar/gkn923
- Ibelli, A., Ribeiro, A., Giglioti, R., Regitano, L., Alencar, M., Chagas, A., … Oliveira, M. (2012). Resistance of cattle of various genetic groups to the tick Rhipicephalus microplus and the relationship with coat traits. Veterinary Parasitology, 186(3), 425–430. doi:10.1016/j.vetpar.2011.11.019
- Jenkins, G. (2003). The emerging role for sphingolipids in the eukaryotic heat shock response. Cellular and Molecular Life Sciences, 60(4), 701–710.
- Jensen, P. (2002). The ethology of domestic animals: An introductory text. CABI Publishing, Wallingford, Oxon OX10 8DE,UK.
- Jian, W., Duangjinda, M., Vajrabukka, C., & Katawatin, S. (2014). Differences of skin morphology in Bos indicus, Bos taurus, and their crossbreds. International Journal of Biometeorology, 58(6), 1087–1094. doi:10.1007/s00484-013-0700-9
- Jin, Y., Zhou, T., Geng, X., Liu, S., Chen, A., Yao, J., … Liu, Z. (2017). A genome‐wide association study of heat stress‐associated SNPs in catfish. Animal Genetics, 48(2), 233–236. doi:10.1111/age.12482
- Kamprath, K., Plendl, W., Marsicano, G., Deussing, J., Wurst, W., Lutz, B., & Wotjak, C. (2009). Endocannabinoids mediate acute fear adaptation via glutamatergic neurons independently of corticotropin‐releasing hormone signaling. Genes, Brain and Behavior, 8(2), 203–211. doi:10.1111/j.1601-183X.2008.00463.x
- Kim, E., Elbeltagy, A., Aboul-Naga, A., Rischkowsky, B., Sayre, B., Mwacharo, J., & Rothschild, M. (2016). Multiple genomic signatures of selection in goats and sheep indigenous to a hot arid environment. Heredity, 116(3), 255–264. doi:10.1038/hdy.2015.94
- Kim, J., Cho, S., Caetano-Anolles, K., Kim, H., & Ryu, Y.-C. (2015). Genome-wide detection and characterization of positive selection in Korean Native Black Pig from Jeju Island. BMC Genetics, 16(1), 1. doi:10.1186/1471-2156-16-S2-S4
- Kim, J., Hanotte, O., Mwai, O. A., Dessie, T., Bashir, S., Diallo, B., … Kim, H. (2017). The genome landscape of indigenous African cattle. Genome Biology, 18(34). doi:10.1186/s13059-017-1153-y
- Kongsuwan, K., Josh, P., Colgrave, M. L., Bagnall, N. H., Gough, J., Burns, B., & Pearson, R. (2010). Activation of several key components of the epidermal differentiation pathway in cattle following infestation with the cattle tick, Rhipicephalus (Boophilus) microplus. International Journal for Parasitology, 40(4), 499–507. doi:10.1016/j.ijpara.2009.10.013
- Kongsuwan, K., Piper, E., Bagnall, N., Ryan, K., Moolhuijzen, P., Bellgard, M., … Jonsson, N. N. (2008). Identification of genes involved with tick infestation in Bos taurus and Bos indicus. Developmental Biology, 132, 77–88.
- Langmead, B., & Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nature Methods, 9(4), 357–359. doi:10.1038/nmeth.1923
- Lee, H.-J., Kim, J., Lee, T., Son, J. K., Yoon, H.-B., Baek, K.-S., … Yang, B.-C. (2014). Deciphering the genetic blueprint behind Holstein milk proteins and production. Genome Biology and Evolution, 6(6), 1366–1374. doi:10.1093/gbe/evu102
- Lee, T.-H., Guo, H., Wang, X., Kim, C., & Paterson, A. H. (2014). SNPhylo: A pipeline to construct a phylogenetic tree from huge SNP data. BMC Genomics, 15(1), 1. doi:10.1186/1471-2164-15-162
- Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., … Durbin, R. (2009). The sequence alignment/map format and SAMtools. Bioinformatics (Oxford, England), 25(16), 2078–2079. doi:10.1093/bioinformatics/btp352
- Low, A. (1990). Nutritional regulation of gastric secretion, digestion and emptying. Nutrition Research Reviews, 3(01), 229–252. doi:10.1079/NRR19900014
- Makina, S. O., Muchadeyi, F. C., van Marle-Köster, E., Taylor, J. F., Makgahlela, M. L., & Maiwashe, A. (2015). Genome-wide scan for selection signatures in six cattle breeds in South Africa. Genetics Selection Evolution, 47(1), 1–14. doi:10.1186/s12711-015-0173-x
- Mapholi, N. O., Marufu, M. C., Maiwashe, A., Banga, C. B., Muchenje, V., MacNeil, M. D., … Dzama, K. (2014). Towards a genomics approach to tick (Acari: Ixodidae) control in cattle: A review. Ticks and Tick-Borne Diseases, 5(5), 475–483. doi:10.1016/j.ttbdis.2014.04.006
- Martinez, M., Machado, M., Nascimento, C., Silva, M., Teodoro, R., Furlong, J., … Azevedo, A. (2006). Association of BoLA-DRB3. 2 alleles with tick (Boophilus microplus) resistance in cattle. Genetics and Molecular Research : GMR, 5(3), 513–524.
- Martinsen, T. C., Bergh, K., & Waldum, H. L. (2005). Gastric juice: A barrier against infectious diseases. Basic & Clinical Pharmacology & Toxicology, 96(2), 94–102. doi:10.1111/j.1742-7843.2005.pto960202.x
- Marufu, M. C., Qokweni, L., Chimonyo, M., & Dzama, K. (2011). Relationships between tick counts and coat characteristics in Nguni and Bonsmara cattle reared on semiarid rangelands in South Africa. Ticks and Tick-Borne Diseases, 2(3), 172–177. doi:10.1016/j.ttbdis.2011.07.001
- McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., … Daly, M. (2010). The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research, 20(9), 1297–1303. doi:10.1101/gr.107524.110
- McManus, C., Do Prado Paim, T., de Melo, C. B., Brasil, B. S., & Paiva, S. R. (2014). Selection methods for resistance to and tolerance of helminths in livestock. Parasite (Paris, France), 21(56), 56. doi:10.1051/parasite/2014055
- Mirkena, T., Duguma, G., Haile, A., Tibbo, M., Okeyo, A., Wurzinger, M., & Sölkner, J. (2010). Genetics of adaptation in domestic farm animals: A review. Livestock Science, 132(1), 1–12. doi:10.1016/j.livsci.2010.05.003
- Mwai, O., Hanotte, O., Kwon, Y.-J., & Cho, S. (2015). African indigenous cattle: Unique genetic resources in a rapidly changing world. Asian-Australasian Journal of Animal Sciences, 28(7), 911–921. doi:10.5713/ajas.15.0002R
- Nakamura, Y., Kanemarum, K., & Fukami, K. (2013). Physiological functions of phospholipase Cδ1 and phospholipase Cδ3. Advances in Biological Regulation, 53(3), 356–362. doi:10.1016/j.jbior.2013.07.003
- Neto, L. R. P., Jonsson, N. N., Michael, J., & Barendse, W. (2011). Molecular genetic approaches for identifying the basis of variation in resistance to tick infestation in cattle. Veterinary Parasitology, 180(3–4), 165–172. doi:10.1016/j.vetpar.2011.05.048
- Nikulina, E. (1990). The brain catecholamines during domestication of the silver fox Vulpes fulvus. Zhurnal Evoliutsionnoi Biokhimii I Fiziologii, 26(2), 156.
- Noyes, H., Brass, A., Obara, I., Anderson, S., Archibald, A. L., Bradley, D. G., … Gicheru, M. (2011). Genetic and expression analysis of cattle identifies candidate genes in pathways responding to Trypanosoma congolense infection. Proceedings of the National Academy of Sciences, 108(22), 9304–9309. doi:10.1073/pnas.1013486108
- Philipp, U., Hamann, H., Mecklenburg, L., Nishino, S., Mignot, E., Günzel-Apel, A.-R., … Leeb, T. (2005). Polymorphisms within the canine MLPH gene are associated with dilute coat color in dogs. BMC Genetics, 6(1), 1. doi:10.1186/1471-2156-6-S1-S125
- Pickrell, J. K., Coop, G., Novembre, J., Kudaravalli, S., Li, J. Z., Absher, D., … Feldman, M. W. (2009). Signals of recent positive selection in a worldwide sample of human populations. Genome Research, 19(5), 826–837. doi:10.1101/gr.087577.108
- Piper, E. K., Jackson, L. A., Bagnall, N. H., Kongsuwan, K. K., Lew, A. E., & Jonsson, N. N. (2008). Gene expression in the skin of Bos taurus and Bos indicus cattle infested with the cattle tick, Rhipicephalus (Boophilus) microplus. Veterinary Immunology and Immunopathology, 126(1), 110–119. doi:10.1016/j.vetimm.2008.06.011
- Price, A. L., Patterson, N. J., Plenge, R. M., Weinblatt, M. E., Shadick, N. A., & Reich, D. (2006). Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics, 38(8), 904–909. doi:10.1038/ng1847
- Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A., Bender, D., … Daly, M. J. (2007). PLINK: A tool set for whole-genome association and population-based linkage analyses. The American Journal of Human Genetics, 81(3), 559–575. doi:10.1086/519795
- Rege, J. (1999). The state of African cattle genetic resources I. Classification framework and identification of threatened and extinct breeds. Animal Genetic Resources Information, 25, 1–25. doi:10.1017/S1014233900003448
- Roth, Z., & Hansen, P. (2004). Sphingosine 1-phosphate protects bovine oocytes from heat shock during maturation. Biology of Reproduction, 71(6), 2072–2078. doi:10.1095/biolreprod.104.031989
- Sabeti, P. C., Varilly, P., Fry, B., Lohmueller, J., Hostetter, E., Cotsapas, C., … Gaudet, R. (2007). Genome-wide detection and characterization of positive selection in human populations. Nature, 449, 913–918. doi:10.1038/nature06250
- Shyma, K., Gupta, J. P., & Singh, V. (2015). Breeding strategies for tick resistance in tropical cattle: A sustainable approach for tick control. Journal of Parasitic Diseases, 39(1), 1–6. doi:10.1007/s12639-013-0294-5
- Sugimoto, N., Shido, O., Matsuzaki, K., Ohno-Shosaku, T., Hitomi, Y., Tanaka, M., … Masaki, Y. (2012). Cellular heat acclimation regulates cell growth, cell morphology, mitogen-activated protein kinase activation, and expression of aquaporins in mouse fibroblast cells. Cellular Physiology and Biochemistry, 30(2), 450–457. doi:10.1159/000339038
- Wang, M., Dzama, K., Rees, D., & Muchadeyi, F. (2015). Tropically adapted cattle of Africa: Perspectives on potential role of copy number variations. Animal Genetics, 47, 154–164. doi:10.1111/age.12391
- Wang, Y., Reverter, A., Kemp, D., McWilliam, S., Ingham, A., Davis, C., … Lehnert, S. (2007). Gene expression profiling of Hereford Shorthorn cattle following challenge with Boophilus microplus tick larvae. Animal Production Science, 47(12), 1397–1407. doi:10.1071/EA07012
- Warwick, E., & Cobb, E. (1975). Genetic Variation in Nutrition of Cattle for Meat Production. Paper presented at the The Effect of Genetic Variance on Nutritional Requirements of Animals, Washington, D.C.
- Wikel, S. K. (1996). Host immunity to ticks. Annual Review of Entomology, 41(1), 1–22. doi:10.1146/annurev.en.41.010196.000245
- Yoshimura, Y. (1997). The ovarian renin–Angiotensin system in reproductive physiology. Frontiers in Neuroendocrinology, 18(3), 247–291. doi:10.1006/frne.1997.0152
- Zeder, M. A. (2012). The domestication of animals. Journal of Anthropological Research, 68(2), 161–190. doi:10.3998/jar.0521004.0068.201
- Zhang, L., Li, W.-H., Anthonavage, M., & Eisinger, M. (2006). Melanocortin-5 receptor: A marker of human sebocyte differentiation. Peptides, 27(2), 413–420. doi:10.1016/j.peptides.2005.05.030
- Zhang, L., Li, W.-H., Anthonavage, M., Pappas, A., Rossetti, D., Cavender, D., … Eisinger, M. (2011). Melanocortin-5 receptor and sebogenesis. European Journal of Pharmacology, 660(1), 202–206. doi:10.1016/j.ejphar.2010.10.100
