?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Enset (Ensete ventricosum) bacterial wilt (EBW) incited by Xanthomonas campestris pv. musacearum (Xcm) is threatening enset production in southwestern Ethiopia. The objectives of this study were to determine the pathogenicity of Xcm isolates and select enset clones resistant to pathogenic Xcm isolates in the study areas. A total of 30 Xcm isolates were subjected to pathogenicity tests on a susceptible enset clone Yeko and all were found pathogenic. Out of 30 pathogenic isolates, three isolates representing three altitude groups [lowland (1470 m.a.s.l), midland (1938 m.a.s.l) and highland (2360 m.a.s.l)] were used for enset clonal evaluation trials. In the clone evaluation trial, 15 enset clones (13 local and a tolerant and susceptible check) were evaluated for 2 years (2017 and 2018) under screen house conditions at Tepi National Spice Research Center, southwestern Ethiopia. The experiments were factorially arranged in a completely randomized design with three replications. An aliquot of 10 ml of the bacterial cell suspension with a concentration of 1 × 108 cfu/ml was inoculated into the second innermost leaf petiole of enset using a sterile hypodermic syringe. Starting from 15 days after inoculation (DAI), data were collected on incubation period (IP), disease incidence (DI), percentage severity index (PSI), days to complete wilting/death (DD), area under disease progress curve (AUDPC) and disease progress rate. Analysis of variance for IP, DI, DD and AUDPC revealed significant differences (P < 0.05) among tested enset clones, while for PSI significant differences (P < 0.05) existed among the interaction effect of enset clones x bacterial isolates. Disease incidence recorded ranges from 0% to 90% and IP ranges from 0 to 23 days. Similarly, the days to complete wilting of susceptible clone reaches up to 63 days, while the calculated AUDPC values ranged from 0 (Gudiro, Maziya and Nobo) to 3190%-days (Arkia, Ataro, Yeko, Chikaro and Ogisso). Disease progress rates also ranged from −0.00165 to 0.04398 units day−1. Clones Gudiro, Maziya and Nobo showed a resistant/tolerant reaction to EBW, while clones Arkia, Ataro, Yeko, Chikaro and Ogisso were the most susceptible enset clones. Based on the results, it is recommended that more clones be evaluated across different agro-ecological areas to select enset clones with stable resistance against the disease. In addition, clone selection should also be given due attention to adaptability as well as quantitative and qualitative yield traits to improve adoption.
PUBLIC INTEREST STATEMENT
Xanthomonas campestris pv. musacearum (Xcm) is a causal agent of enset (Ensete ventricosum) bacterial wilt (EBW) in Ethiopia and banana Xanthomonas wilt in Africa. Nowadays, this pathogen is diminishing foods obtain from enset (kocho, bulla and amicho) and banana fruit by totally killing the plants. The EBW has also forced farmers to abandon enset production (more than 20 million populations in Ethiopia were deepened on enset), resulting in critical food shortage in the densely populated areas of southern Ethiopia. Therefore, determining the pathogenicity of the Xcm isolates and evaluating local enset clones for their resistant to pathogenic Xcm isolates could help to come up with some resistant enset clones to control the EBW caused by this pathogen. Hence, by so doing, three enset clones, namely, Gudiro, Nobo and Maziya were found resistant for Xcm and can be used on integrated EBW management system at southwestern Ethiopia and similar agro-ecologies.
Competing Interests
The authors declare no competing interests.
1. Introduction
In Ethiopia, enset (Ensete ventricosum (Welw.) Cheesman) is an important staple and co-stable crop grown for food, feed, medicinal, cultural and industrial purposes (Ajebu et al., Citation2008; Yemane & Fassil, Citation2006). The crop is mainly grown in the southern and southwestern parts of the country (Bezuneh & Feleke, Citation1966), and is the main food source for over 20 million people in the region (Zerihun et al., Citation2013). The edible parts of enset are the pseudostem and corm in the form of kocho, bulla and amicho. Kocho is the fermented starch obtained from decorticated leaf sheaths and grated corms. Bulla is obtained by squeezing out the liquid containing starch from scraped leaf sheaths and grated corm allowing the starch to concentrate into a white powder. Amicho is a piece of enset corm that is boiled and consumed in a similar manner to other root and tuber crops. Moreover, enset plants are used for fiber production, which is used for various cultural and industrial activities (Almaz & Anke, Citation2004; Brandt et al., Citation1997; Endale et al., Citation1994).
Despite its importance, enset is constrained by several biotic and abiotic factors that affect its production and productivity. Among the biotic factors, diseases, insect pests and wild animals are important production challenges of enset. But, of all, enset bacterial wilt (EBW) caused by Xanthomonas ampestris pv. musacearum (Xcm) is the most damaging constraint contributing a major share to reduction of enset productivity in all enset growing areas of Ethiopia (Fikre & Alemar, Citation2016; Mekuria et al., Citation2016). Recent work based primarily on DNA sequence and fatty acid data has shown that strains of Xcm have very close homology to strains of Xanthomonas vasicola and most likely belong to this species. Accordingly, the name X. vasicola has been proposed for Xcm (Aritua et al., Citation2008; Studholme et al., Citation2020), though this has not been formally approved as a new combination of names by the International Committee on Systematic Bacteriology. Therefore, the previous name Xcm is still the current official name. The pathogen Xcm, invades the vascular system of enset, causing wilting (yellowing) of the heart-leaf, followed by progressive yellowing and necrosis of the neighboring overlapping leaves, and eventual death of the plant. During the opening of wilted petioles, yellow or cream colored bacterial oozes are clearly observed (Gizachew, Citation2000).
In Ethiopia, an EBW epidemic was reported by Dereje (Citation1985) with losses up to 70%. Recently, many researchers reported losses of up to 100% in some enset fields in southwestern Ethiopia due to EBW leading to a continuous declining of the area and productivity of enset (Desalegn & Addis, Citation2015; Mekuria et al., Citation2016; Tariku et al., Citation2015). Other than Ethiopia, the disease occurs in banana-growing areas of Africa, such as Uganda (Tushemereirwe et al., Citation2003), the Democratic Republic of Congo (Ndungo et al., Citation2005), Rwanda (Reeder et al., Citation2007), Kenya, Tanzania and Burundi (Carter et al., Citation2010). The situation calls for developing designing of effective EBW management options to sustain the productivity of the crop.
However, control of EBW is a real challenge as there are no clones with known resistance, commercially available chemicals or biocontrol agents. Although enormous advisory services on field sanitation (disinfection of farming and processing tools, and rouging of infected enset plants) have been undertaken to manage the disease, the strategy was reported difficult to implement by smallholder farmers. Host plant resistance is believed to be the most effective and economical management option for EBW. Reports related to clonal screening against bacterial wilt have indicated the possibilities of managing EBW through host plant resistance (Mengistu et al., Citation2014). Evaluation and identification of resistant/tolerant enset clones to EBW and availing host resistance to the farmer contributes towards managing the disease in a sustainable manner. Therefore, the objectives of this research were (1) to elucidate the pathogenicity of Xcm isolates, and (2) to evaluate the resistance reaction of selected enset clones to Xcm isolates collected from southwestern Ethiopia.
2. Materials and methods
2.1. Area description
Enset bacterial wilt was studied in three administrative zones, namely, Bench-maji, Keffa and Sheka of southwestern Ethiopia. Southwestern Ethiopia is characterized by humid and tropical rainforest climate type and forest coffee systems. The altitude of study areas ranged from 1470 to 2393 m.a.s.l and is located within 4.43°-8.58°N latitude and 34.88°-39.14°E longitude.
The study area receives an average annual rainfall between 1800 and 2200 mm with multimodal distribution and experiences annual mean temperature ranging between 15.1°C and 27°C (Bedru, Citation2007). The dominant crops grown by farmers are enset (Enseteventricosum), potato (Solanumtuberosum), wheat (TriticumaestivumL.), maize (Zea mays L.), barley (Hordeum vulgare), teff (Eragrostistef), coffee, taro and others.
2.2. Sample collection
Enset bacterial wilt samples were collected from wilted enset plant leaf petioles during the main rainy season of the 2017 cropping year. Leaf petioles showing early stage of the disease symptom were collected to avoid some saprophytic microorganisms that grow in tissues killed by the primary pathogen. Based on enset growing potential as well as available budget and time resources, a total of 120 representative samples from 30 Farmer Associations (FAs) in 10 districts of southwestern Ethiopia were collected. Bacteria cells oozing out of the vascular tissues of infected enset plants were randomly taken using sterile toothpick and then suspended in sterilized distilled water in half-filled screw-capped vials according to Quimio (Citation1992). Each sample was labeled with location (zone, district and FAs) and altitude ranges.
2.3. Isolation, identification and preservation of the pathogen
From each collected isolate, a loopful of the suspension was streaked on yeast peptone sucrose agar (YPSA) medium, which composed of yeast extract, 5 g; peptone, 10 g; sucrose, 20 g; agar, 12–15 g in 1 L distilled water with pH 7.4 and autoclaved at 121°C for 15 minutes. The plates were incubated at 28°C for 48–72 hours as suggested by Schaad and Stall (Citation1988). After 2–3 days of incubation, bacterial colonies from each plate were further sub-cultured to develop pure cultures of the isolates and transferred to YPSA slants incubated at 28°C for 48–72 hours. The isolates were characterized and identified based on the cultural, morphological and a biochemical description in Bergey’s manual of systematic bacteriology (Bergey, Citation1930). Pure cultures were preserved at 4°C for further successive activities in Plant Pathology Laboratory of Haramaya University, Ethiopia.
2.4. Pathogenicity test
2.4.1. Growing susceptible clone
A recommended local susceptible enset clone Yeko was used for pathogenicity test (Befekadu et al., Citation2014). One-year-old suckers of the clone were transplanted to pots (30 × 30 cm), filled with a sterilized mixture of topsoil, composted manure and sand with 3:2:1 ratio (Quimio, Citation1992), then allowed to establish for 3 months in screen house (with 20 m length and 12 m width) at 25–30°C day and 15–18°C night temperatures at Tepi National Spice Research Center (TNSRC). Plants were watered uniformly every day both in the morning and in the afternoon to maintain the relative humidity within the range of 60% to 80% up to the end of the assessment periods.
2.4.2. Inoculum preparation and inoculation
From identified isolates, 30 Xcm isolates were selected (single isolate from each cluster/FAs) and subjected to pathogenicity test. The selected isolates were grown on YPSA medium at 28°C for 48 hours to prepare a sufficient amount of inocula. Then, bacterial cells were independently harvested by scratching and suspending the culture in sterilized distilled water in a sterile beaker and adjusted to a concentration of 1 × 108 cfu/ml using a spectrophotometer (Fikre & Gizachew, Citation2007). Three milliliters of the bacterial cell suspension were inoculated using a sterile hypodermic syringe to the second innermost leaf petiole of a 3-month-old enset plant. Three plants were used as replica and sterilized distilled water-inoculated plants were included as negative control.
2.5. Clonal evaluation
2.5.1. Enset clones
Fifteen enset clones, namely, Ataro, Baradi, Barasho, Boso, Chikaro, Gudiro, Kekaro, Nobo, Ogisso, Shisho, Tafaro, Utiro and Yeko (local clones); and Arkia and Meziya (recommended susceptible and tolerant check, respectively) were used; and they were described and differentiated based on their morphological characters following the procedures in Tesfaye (Citation2008) (Table ). The enset clones Arkyia and Meziya were collected from maintenance plots of Areka Agricultural Research Centre (AARC), Ethiopia. The other clones were collected from farmers’ fields in southwestern Ethiopia, where these clones are commonly grown. In southwestern Ethiopia, farmers differentiate one enset clone from the other phenotypically by looking at the colour of the petiole, leaf sheath, angle of leaf orientation, size and colour of leaves and pseudostem. This mode of classification is also well reported and documented in the literature (Tesfaye, Citation2008).
Table 1. Description of 15 enset clones evaluated for resistance reaction to pathogenic Xanthomonas campestris pv. musacearum isolates collected from southwestern Ethiopia
2.5.2. Inoculum selection and preparation for inoculation
Based on their short incubation period and date to complete wilting recorded during the pathogenicity test, 10 Xcm isolates, namely Xcm-7, Xcm-20, Xcm-26, Xcm-43, Xcm-49, Xcm-69, Xcm-73, Xcm-93, Xcm-108 and Xcm-117 were selected and subjected to test their ability to induce a hypersensitivity reaction on tobacco (Nicotiana tabacum) leaves based on the method described by Fikre and Zeller (Citation2007). Finally, three bacterial isolates were selected and upgraded for clonal evaluation trial, based on their ability to infect a non-host tobacco plant aggressively within 72 hours. These isolates represented different agro-ecologies. That is, Xcm-L (Xcm-117), Xcm-M (Xcm-7) and Xcm-H (Xcm-49) as they were originated from lowland (1470 m.a.s.l), midland (1938 m.a.s.l) and highland (2360 m.a.s.l) areas, respectively.
The three Xcm isolates were grown on YPSA medium at 28°C for 48 hours and bacterial cells were prepared for inoculation in a similar manner to the pathogenicity test in Section 2.3.2.
2.5.3. Experimental design
The treatments were arranged in 4 × 15 factorial experiments in a completely randomized design with three replications. The factors were three Xcm isolates and control (sterilized distilled water) and 15 enset clones. Thus, a total of 180 one-year-old enset suckers were planted in pots (single plant per pot), and inoculation was made after the plants well established (7 months after planting). An aliquot of 10 ml of the bacterial cell suspension was inoculated using a sterile hypodermic syringe to the second innermost leaf petiole of enset plant; and the inoculated wounds were sealed with waterproof sticky tape. The control plants were also inoculated with the same amount of sterilized distilled water. The temperature of the screen house was recorded as 25–30°C in the day and 15–18 oC in the night; and the relative humidity was 60% to 80% during the experimental periods. The typical wilting symptom development (yellowing and necrosis of leaves) was observed and recorded on test plants. The experiment was done twice. The first round of the experiment was conducted from 30 April 2017 to 7 January 2018, and the second experiment was executed from 5 February 2018 to 9 October 2018.
2.6. Data collection
From pathogenicity test, starting from first date of symptom development, data on EBW typical wilting symptom, incubation period (a period between inoculation and first wilting symptom, IP) and days to complete wilting/death (a period between inoculation and complete wilting, DD) were observed and recorded. Finally, the bacterial oozes from infected leave petioles were aseptically collected, re-isolated and compared with the respective parent colony characteristics. Moreover, from clonal evaluation experiment, data on EBW incubation period, disease incidence (DI), and days to complete wilting/death were also recorded. Disease incidence was recorded six times at 10-day interval (Fikre & Alemar, Citation2016) for 65 days. Enset bacterial wilt incidence was determined as mean percentage of enset plants showing typical wilt disease symptoms (yellowing and necrosis of leaves) of inoculated enset clones using the following formula:
As some enset clones were recovering after infection, disease severity was also assessed by using a 0–5 disease scoring scale; where 0 = no visible disease symptom, 1 = 1 leaf wilted, 2 = 2–3 leaves wilted, 3 = 4 leaves wilted, 4 = all leaves wilted, and 5 = the plant has dead (Winstead & Kelman, Citation1952). Disease severity scores were converted into percentage severity index (PSI) for analysis (Wheeler, Citation1969) as:
Moreover, area under disease progress curve (AUDPC) was computed from severity data using the formula suggested by Campbell and Madden (Citation1990):
where n = the number of observations, ti = time of the ith disease assessment; and xi = disease severity at ith assessment. As severity was recorded in percent and the time of disease assessment was made days after inoculation, AUDPC was expressed in percent-days (%-days).
2.7. Data analyses
The DI and PSI data were transformed using Arc sine (angular) transformation to stabilize the response variance to the normal distribution. All the collected quantitative data were subjected to analysis of variance (ANOVA) using the SAS software version 9.2 (SAS Institute Inc, Citation2008). Means were compared using Tukey’s test according to Gomez and Gomez (Citation1984). Significant differences between and among means were examined at 5% probability level. To determine the disease progress rate from PSI, a Monomolecular model, ln[1/(1-y)] (Van der Plank, Citation1963) was used to estimate the disease progression. The transformed PSI data were regressed over time (days after inoculation) to determine the disease progress rate. The slope of the regression line estimated the disease progress rate. Regression was computed using Minitab (Release 18.0 for windows®, 2007). The two runs of experiments were pooled because of homogeneity of variances as tested using Bartlett’s test (Gomez & Gomez, Citation1984) and the F-test was not significant for the parameters studied. Thus, data were combined for analysis.
3. Results
3.1. Pathogenicity test
The results of pathogenicity test revealed that all the tested isolates of Xcm infected the susceptible enset clone, yeko, within 12–32 days after inoculation (DAI) (Table ). The inoculated leaves showed light yellow to dark brown necrosis symptoms around the inoculated areas and those leaves then became yellowish brown and finally dried from apex end till the petiole collapsed. These typical symptoms started on the inoculated leaf and spread gradually to the remaining leaves of the plant leading to complete death within 32–55 DAI (Table ). Typical Xcm oozes were observed by opening petioles of symptomatic leaves. These symptoms were consistent with bacterial wilt of enset observed in the field during disease assessment. Re-isolation and identification also confirmed their similarities with the parent isolates. Those leaf parts inoculated with sterilized distilled water remained healthy until the end of the study periods.
Table 2. Comparison of Xanthomonas campestris pv. musacearum isolates for the appearance of first disease symptoms (incubation period) and days for complete wilting after inoculation on enset clone, Yeko, in screen house conditions in southwestern Ethiopia, during 2017
The isolates Xcm-7, Xcm-49 and Xcm-117 had significantly (P < 0.001) lower IP (13, 12 and 12 DAI, respectively) than Xcm-99 with IP of 32 DAI (Table ). The isolates Xcm-7, Xcm-49, Xcm-93, Xcm-103 and Xcm-117 caused a complete wilting (death) of plants within significantly (P < 0.001) shorter times (32–33 DAI) than Xcm-10 that took 55 DAI with the remaining isolates comprising a stratified intermediate (Table ).
3.2. Evaluation of enset clones for resistance reaction to Xcm
All the tested enset clones, with the exception of Maziya and Nobo, started to show typical wilt symptoms within 15–23 days after inoculation. The symptoms developed were similar to those observed in naturally infected enset plants during the field visit. Initial symptoms first appeared on the inoculated leaf petioles in which leaf turned from green to deep yellow (Figure ) as compared to healthy enset leaves (Figure ). These symptoms spread gradually to the remaining leaves, and the infected leaf tips became limp and dropped (Figure ). A cut made through the petioles of a newly infected enset plant revealed browning of the vascular strands and yellowish or grayish masses of bacterial load that oozes out from the strands. Such bacteria oozes were different in color, from light yellow to deep yellow, and from one enset plant to another. Gradually, all the leaves wilted, bended over and withered (Figure ). But these symptoms were not observed on the clone Maziya and Nobo as well as on the control.
Figure 1. Inoculated enset clones showing no and variable wilt symptoms. (a) healthy enset clones; (b) enset bacterial wilt symptom appearing on first inoculated leaf petiole after artificially inoculated with Xanthomonas campestris pv. musacearum; (c) disease progressed to the remaining leaf petioles and yellowing of leaves; (d) mixture of tolerant/resistant enset clones and completely wilted (died) enset clone.

There were very highly significant (P < 0.0001) differences among the tested enset clones in all the disease parameters considered (Table ). But, with the exception of PSI, the interaction effects of enset clones x bacterial isolates were not significant (Table ). Enset clones Arkia, Ataro, Baradi, Barasho, Boso, Chikaro, Kekaro, Ogisso, Shisho, Tafaro, Utiro and Yeko showed significantly (P < 0.05) higher mean disease incidence ranging from 81% to 90% with IP ranging from 15 to 23 days (Table and Appendix B Tables 1 and 2). On the contrary, no disease recorded on Gudiro, Maziya and Nobo enset clones evaluated, but clone Gudiro showed disease symptoms on inoculated leaf petioles after 20 days of inoculation (Table ).
Table 3. Mean squares of 15 enset (Ensete ventricosum) clones evaluated against three pathogenic Xanthomonas campestris pv. musacearum isolates and associated disease parameters under screen house conditions in southwestern Ethiopia, during 2017/18
Table 4. Mean incubation period, disease incidence, days to complete wilting or death and area under disease progress curve of 15 enset clones evaluated against three pathogenic Xanthomonas campestris pv. musacearum isolates under screen house conditions in southwestern Ethiopia, during 2017/18
Almost all the tested enset clones, except Gudiro, Maziya and Nobo, wilted completely after infection with Xcm isolates. The period between inoculation and complete wilting (DD) varied significantly (P < 0.05) between enset clones (Tables , and Appendix B Table 3). Inoculated plants of enset clone Gudiro, Maziya and Nobo did not wilt completely. Plants of the clone Gudiro, which showed the disease symptom on its inoculated leaf petioles recovered from the disease within 55 days after inoculation. And eventually these recovered plants resembled healthy control plants. In contrast, enset clones Arkia and Ataro wilted completely earlier within 48 and 54 days after inoculation, respectively, than clones Baradi, Barasho, Chikaro, Kekaro, Ogisso, Shisho, Utiro and Yeko, which showed complete wilting within 57 to 63 days after inoculation. But enset clones Boso and Tafaro wilted after relatively longer periods of time, 63 days after inoculation, on average (Table ).
Enset clones Arkia, Ataro, Yeko, Chikaro and Ogisso showed significantly (P < 0.05) high mean AUDPC values of 3190, 2920, 2869, 2690 and 2572%-days, respectively. On the other hand, significantly (P < 0.05) low AUDPC values were recorded from enset clones Gudiro, Maziya and Nobo (Table and Appendix B Table 4). Enset clones Arkia (90%), Chikaro (90%), Ataro (88%) and Yeko (88%) showed significantly (P < 0.05) higher mean disease severity than enset clone Barasho (64%) and Baradi (61%) (Table and Appendix B Table 5). But, no disease severity recorded from enset clones Gudiro, Maziya and Nobo. The enset clone Baradi had a differential resistance reaction to the three bacterial isolates tested. This clone was highly infected by isolate Xcm-H and Xcm-M with respective severity of 78% and 72%, and the former isolate was significantly (P < 0.05) different from the later isolate with moderate PSI value of 32% (Table ).
Table 5. Final percentage severity index of 15 enset clones evaluated against three pathogenic Xanthomonas campestris pv. musacearum isolates under screen house conditions in southwestern Ethiopia, during 2017/18
3.3. Disease progress rate (r) and curves
Disease progress rates on the 15 enset clones tested ranged from—0.00165 to 0.04398 units day−1 (Table ). The disease progress rate was relatively higher on Chikaro × Xcm-L, Arekia × Xcm-L, Ataro × Xcm-L, Yeko × Xcm-M, Ataro × Xcm-M, Yeko × Xcm-L and Arekia × Xcm-M than other host-isolate combinations studied. On the other hand, lower disease progress rate was recorded on Baradi × Xcm-L, Barasho × Xcm-L, Boso × Xcm-L, Barasho × Xcm-H, Barasho × Xcm-M, Boso × Xcm-M and Utiro × Xcm-M combinations than the rest. There was no disease severity recorded from enset clones Nobo and Maziya, and hence, the disease progress rates were zero. Rates computed from enset clone Gudiro × Xcm-H, Xcm-M and Xcm-L were −0.00165, −0.00143 and −0.00024 units day−1, respectively, implying for degressive nature of the disease on the clone tested. This could confirm that the plant became healthy late in the assessment period due to recovering from the infection. The rate values of enset clone Nobo, Maziya and control are not presented in Table , as zero PSI values were recorded throughout the assessment period for both clones.
Table 6. Mean initial (PSIi) and final (PSIf) severity index and parameter estimates of enset bacterial wilt (Xanthomonas campestris pv. musacearum) on enset clones in response to Xcm inoculation under screen house conditions in southwestern Ethiopia, during 2017/18
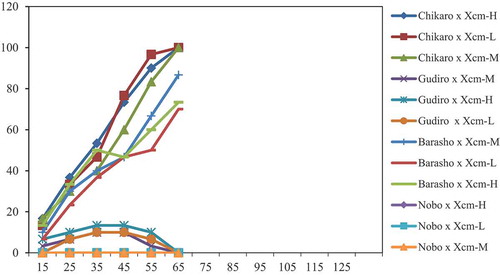
Similarly, the disease progress curves (severity versus DAI) sketched from PSI data showed differences for each enset clone and Xcm isolate combinations. The disease progress curves in Figure depicted only for the combinations of four enset clones (Chikaro, Gudiro, Barasho and Nobo) and the three Xcm isolates. These combinations were sketched based on EBW severity levels compared with susceptible and tolerant clones for the sake of clarity and ease of graphic presentation. Accordingly, severity in enset clone Chikaro experienced a relatively highly raised progressive curve and displayed the highest level of PSI. Whereas, disease progress curves of enset clone Nobo showed no progression and lied on X-axis for each isolate combination. And, disease progress curves of enset clone Gudiro x Xcm isolates progressed slowly and degressed rapidly and declined to the X-axis in response to recovery of infection late in the epidemic period.
4. Discussion
Enset bacterial wilt, caused by Xcm, is considered as one of the major biotic stresses threatening enset production. According to Tariku et al. (Citation2015) and Mekuria et al. (Citation2016), initial symptoms of EBW appear on the central heart leaf or on one of the inner leaves of its tip and became yellowish, limp and droop. A cut made through the petioles of a newly infected enset plant reveals browning of the vascular strands and yellowish or grayish masses of bacteria ooze out from the strand. Cross sections at the base of pseudostem and corm show discoloration of the vascular strand with large bacterial pocket while grayish or yellowish exudates with brownish to black spot were observed from corm cross sections in the study. Moreover, Gizachew et al. (Citation2008) further observed, in a more advanced stage of disease development, most of the leaves wilt, breaks at the petiole and wither. Eventually, the whole plant dies and rots to the ground. In the present study, both in pathogenicity and clonal evaluation tests, a similar phenomenon was observed on most of the enset clones studied.
Results of the current pathogenicity tests confirmed that all the tested bacterial isolates were pathogenic to the susceptible enset clone Yeko. Isolates Xcm-7, Xcm-49, Xcm-93, Xcm-103 and Xcm-117 which had relatively lower days of IP and DD were considered as highly pathogenic. In contrast to our findings, Kidist (Citation2003) reported the failure of Xcm isolates in symptom development during pathogenicity test, and the author concluded that the possible reason for failure in pathogenicity could be the effect of the growth media (YPSA). Although YPSA was recommended for all Xanthomonas campestris pathovars, there could be mutation or loss of virulence of the bacterial isolates while growing on this medium.
During clonal evaluation, EBW symptoms were observed in inoculated leaf of enset clone Gudiro and these symptoms did not successfully progressed to neighboring leaf petioles, and inoculated plants appeared healthy thereafter. This could be possible because the bacteria might stay confined to the leaf petiole and leaf sheath of this inoculated leaf. Probably the bacteria could not enter the corm and hence cannot infect adjacent leaves as the vascular connection between leaves passes through the corm. Possibly this is due to the hypersensitive reaction of enset clone. According to Gizachew et al. (Citation2008), this apparent recovery may be explained by the un-systemic nature of the disease development after an artificial inoculation in the leaf petiole of a newly formed leaf. In addition, hypersensitive reactions, which are characterized by the rapid death of individual plant cells due to pathogenic bacteria, could have also contributed to the recovery of resistant plants as commented by Kiraly (Citation1980). This would result in the disappearance of the disease when the inoculated leaf eventually wilts and dies. Fikre and Alemar (Citation2016) also observed that artificially inoculated enset clones Hella, Dirbo, Wachiso, Sirariya, Falakia, and Gisiro recovered from Xcm infection with a related analogy.
The present study revealed significant differences among enset clones. However, no significant difference among Xcm isolates and their interaction with enset clones for disease parameters. Such results confirmed the existence of variation in resistance in the host. Enset clones Gudiro, Maziya and Nobo showed a high level of resistance to Xcm isolates where the isolates did not show pathogenicity the clones. All the Xcm isolates were pathogenic to enset clones Arkia, Ataro, Barasho, Boso, Chikaro, Kekaro, Ogisso, Shisho, Tafaro, Utiro and Yeko, but the isolates differ in their pathogenicity to enset clone Baradi. Similarly, Fikre and Alemar (Citation2016) reported that insignificant bacterial wilt disease infection on artificially inoculated enset clones Alagena, Godere, Meziya, Nechuwe, Nobo, Tikur enset, Unjeme and Yesha were recorded, which were collected from different parts of southern Ethiopia. The authors also reported the susceptibility of enset clones Aganiye, Arkia, Birbo, Ginbo,Gosalo, Kokari, Shena, Tida, Tsella to Xcm infections in a controlled setup.
All the tested enset clones, except Maziya and Nobo, developed disease symptoms between 15 and 23 days after inoculation. It could be a confirmation of the pathogenicity of Xcm isolates that can be performed and proved within 15–23 days by using the susceptible enset clone. Tripathi et al. (Citation2008) also found that after artificial infection with Xcm isolates, no plants of banana cultivar Musa balbisiana wilted completely. In contrast, all the plants of Pisang awak wilted completely within 24 to 28 days after inoculation. In agreement with this, we found that no plants of enset clone Gudiro, Maziya and Nobo wilted completely. On the other hand, all the plants of enset clones Arkia and Ataro wilted completely within a relatively short period of time. And, the clones Boso and Tafaro took longer date to complete wilting after inoculation.
Analysis of AUDPC, PSI and disease progress rate indicated that Gudiro, Maziya and Nobo were the most resistant/tolerant enset clones to EBW, while the enset clones Arkia, Ataro, Chikaro, Kekaro, Ogisso, Shisho, Tafaro, Utiro and Yeko were the most susceptible with the remaining enset clones (Baradi, Barasho and Boso) comprising a stratified intermediate. Enset clones Arkia, Chikaro, Ataro and Yeko were highly infected by all the Xcm isolates, but isolate Xcm-L caused a moderate infection on enset clone Baradi, and all isolates could not infect enset clone Gudiro, Maziya and Nobo. These were probably due to high genetic diversity within enset clones and narrow genetic diversity within Xcm isolates studied. Similarly, Aritua et al. (Citation2008) found that the genetic diversity amongst Xcm strains has been shown to be very narrow, and thus, investigation for host resistance is somewhat simplified in that host/strain interactions are unlikely and any variation in disease expression recorded is most likely to be attributed to the host genotype.
During enset clones collection, we observed that farmers in enset farming communities grow and maintain mixed enset clones in the same field. Unlike other field crops, growing diverse enset clones in the same field is a direct reflection of the various uses of the crop. Diversity in enset clones growing in the inspected areas ensures stable food sources in times of unfavorable environmental conditions and make the availability of wide range of resistance/tolerant to the EBW. At many enset clone collection sites, the farmers commented that EBW is unable to infect the enset clones Gudiro, Maziya and Nobo, and these clones are very strong and hard to withstand some adverse conditions including animal attacks (mole rat and porcupines) to some extent. The production locations of enset clones Gudiro and Nobo are at Sheka zone (Masha and Andiracha districts), while enset clone Maziya is widely grown at Wolayita zone (Areka district) and Dawro zone (Waka district). Therefore, the planting materials of these clones can be obtained from their respective localities. Farmers also suggested that kocho of enset clones Maziya and Nobo are less in quality in terms of colour (dark) as compared to kocho obtained from other clones used in this experiment, which is white in colour. Due to dark colour of kocho, its preference by farmers and market values are expected to be low. Kocho, bulla and fiber obtained from enset clone Gudiro are high in quality and market preference. In general, the yield of enset clones Gudiro, Maziya and Nobo is not inferior, but they are late maturing (>4 years) as compared to the susceptible clones recorded in the study areas.
5. Conclusions
The current work reveals that all of the Xcm isolates are pathogenic to the most of the ecologically adapted enset clones in southwestern Ethiopia. But, Gudiro, Maziya and Nobo have exhibited better resistance reaction to enset bacterial wilt. Hence, Gudiro and Nobo should be considered as resistant/tolerant clones to the pathogen and these materials can be used as a component in enset bacterial wilt management schemes. It is possible to include clone Maziya to enset bacterial wilt management component after taking research on its adaptability to the local agro-ecology. Promising results indicate that resistant genetic material from enset growing regions of southwestern Ethiopia could be identified and developed. Further detailed work involving many isolates and diversified clones should be carried out for a clear understanding of the genetic backgrounds associated with pathogenicity of the Xcm isolates and resistance of enset clones in southwestern Ethiopia.
Acknowledgements
The authors acknowledge the Ministry of Education of Ethiopia and Mizan-Tepi University for financing the study and Haramaya University for providing laboratory facilities during the isolate identification and characterization studies. The authors would also like to thank Tepi National Spice Research Center for facilitating the screen house studies.
Additional information
Funding
Notes on contributors
Befekadu Haile
Mr. Befekadu Haile, is a PhD candidate at Haramaya University, Ethiopia with field of specialization in Plant Pathology. He served as a lecturer and researcher at Mizan-Tepi University, Ethiopia. He has got MSc degree in Plant Pathology from Jimma University, Ethiopia. His primary research interests are population dynamics of plant pathogenic bacteria and studying the temporal and spatial distribution of various plant diseases.
References
- Ajebu, N., Adugna, T., Lars, O., & Frik, S. (2008). Chemical composition and in sacco dry matter degradability of different morphological fractions of 10 enset (Ensete ventricosum) varieties. Animal Feed Science and Technology, 146(2), 55–18.
- Almaz, N., & Anke, N. (2004). The significance of enset culture and biodiversity for rural household food and livelihood security in southwestern Ethiopia. Agriculture and Human Values, 21(1), 61–71. https://doi.org/10.1023/B:AHUM.0000014023.30611.ad
- Aritua, V., Parkinson, N., Thwaites, R., Heeney, J., Jones, D., Tushemereirwe, W., Crozier, J., Reeder, R., Stead, D., & Smith, J. (2008). Characterization of the Xanthomonas sp. Causing wilt of enset and banana and its proposed reclassification as a strain of X. vasicola. Plant Pathology, 57(1), 170–177. https://doi.org/10.1111/j.1365-3059.2007.01687.x
- Bedru, S. (2007). Land cover changes in Andercha and Masha woredas of Sheka Zone SNNP Regional state in forest of sheka.
- Befekadu, H., Grima, A., & Fikire, H. (2014). Physiological characteristics and pathogenicity of Xanthomonas campestris pv. musacearm strain collected from enset and banana in southwest Ethiopia. African Journal of Biotechnology, 13(24), 2425–2434. https://doi.org/10.5897/AJB2014.13794
- Bergey, D. H. (1930). Bergey’s manual of systematic bacteriology. (2nd ed., Vol. II). (Krieg, N. R. and Holt, J.G. eds). Williams and Wilkins.
- Bezuneh, T., & Feleke, A. (1966). The production and utilization of the genus Ensete in Ethiopia. Economic Botany, 20(1), 65–70. https://doi.org/10.1007/BF02861927
- Brandt, S., Spring, A., Hiebsch, C., McCabe, S., Endale, T., Mulugeta, D., Gizachew, W., Gebre, Y., Shigeta, M., & Shiferaw, T. (1997). The ‘tree against hunger’. Enset-based agricul- tural systems in Ethiopia. American Association for the Advancement of Science.
- Campbell, C. L., & Madden, L. V. (1990). Introduction of plant disease epidemiology. John Wiley and Sons.
- Carter, B., Reeder, R., Mgenzi, S., Kinyua, Z., Mbaka, J., Doyle, K., Nakato, V., Mwangi, M., Beed, F., Aritua, V., Lewis-Ivey, M., Miller, S., & Smith, J. (2010). Identification of Xanthomonas vasicola (formerly X. campestris pv. musacearum), causative organism of banana xanthomonas wilt, in Tanzania, Kenya and Burundi. Plant Pathoogy, 59(2), 403. https://doi.org/10.1111/j.1365-3059.2009.02124.x
- Dereje, A. (1985). Studies on the bacterial wilt of enset (Ensete ventricosum) and prospects for its control. Ethiopian Journal of Agricultural Science, 7(1), 1–14.
- Desalegn, R., & Addis, S. (2015). Enset (Ensete ventricosum (Welw.) bacterial wilt survey in Borana Mid-altitude. International Invention Journal of Agricultural and Soil Science, 3(2), 9–12.
- Endale, T., Terefe, B., Mukgeta, D., & Geleta, L. (1994). Improvement study on enset and Sweet potato. In T. Abate, C. Hiebsch, S. A. Brandt, & S. Gebremariam (Eds.), Proceedings of second national horticultural workshop in Ethiopia 1–3 Dec. 1992 (pp. 228-234). The Institute.
- Fikre, H., & Alemar, S. (2016). Enset clones responses to bacterial wilt disease (Xanthomonas campestris pv. Musacearum). International Journal of Applied and Pure Science and Agriculture (IJAPSA), 02(9), 45–53.
- Fikre, H., & Gizachew, W. (2007). Evaluation of enset clone meziya against enset bacterial wilt. African Crop Science Conference Proceedings (Vol. 8, pp. 887–890). African Crop Science Society.
- Fikre, L., & Zeller, W. (2007). Pathogenic characterisation of strains of Ralstonia solanacearum from Ethiopia and influence of plant age on susceptibility of hosts against R. solanacearum. Journal of Plant Diseases and Protection, 114(6), 241–249. https://doi.org/10.1007/BF03356224
- Gizachew, W. (2000). Variation in isolates of enset pathogen (Xanthomonas campestris pv. musacearum) and reaction on enset clones (Ensete ventricosum (Welw) Cheesman to this disease (MSc.Thesis). Alemaya University.
- Gizachew, W., Kidist, B., Blomme, G., Addis, T., & Mengiesha, T. (2008). Evaluation of enset clones against enset bacterial wilt. African Crop Science Journal, 16(1), 89–95.
- Gomez, K. A., & Gomez, A. A. (1984). Statistical procedures for agricultural research. John Wiley and Sons.
- Kidist, B. (2003). Characterization of Xanthomonas campestris pv. musacearum Isolates: Causal agent of enset bacterial wilt disease (MSc.Thesis). Addis Ababa University.
- Kiraly, Z. (1980). Defenses triggered by the invader: Hypersensitivity. In J. G. Horsfall, and F. B. Cowling (Eds.). Plant Disease, 5(1), 201–224.
- Mekuria, W., Amare, A., & Alemayehu, C. (2016). Assessment of bacterial wilt (Xanthomonas campestris pv. musacearum) of enset in Southern Ethiopia. African Journal of Agricult- Ural Research, 11(19), 1724–1733. https://doi.org/10.5897/AJAR2015.9959
- Mengistu, O., Kassahun, S., Tariku, H., & Thangavel, S. (2014). Assessment of disease intensity and evaluation of enset clones against bacterial wilt (Xanthomonas campestris pv. musacearum) in Tikur Inchini and Jibat Districts of West Shewa, Ethiopia. International Journal of Research in Science, 1(2), 83–97. https://doi.org:10.15613/sijrs/2014/v1i2/6754
- Ndungo, V., Bakelana, S., Eden-Green,, & Blomme, G. (2005). An outbreak of banana Xanthomonas wilt in the democratic Republic of Congo. Infomusa, 13(2), 43–44. https://doi.org:10.1111/mpp.12578
- Quimio, J. (1992). Annual report of the plant pathologist 16-17 July 1991. Enset Team Support Project Sidama Gamo Gofa. Peasants Agricultural Development Program PADEPIII. Awasa Research Center (IAR).
- Reeder, R. H., Opolot, O., Muhinyuza, J. B., Aritua, V., Crozier, J., & Smith, J. (2007). Presence of banana bacterial wilt (Xanthomonas campestris pv. musacearum) in Rwanda. New Disease Reports.
- SAS Institute Inc. (2008). SAS/STATA User Guide for Personal Computers Version 9 (2nd ed.). SAS Institute.
- Schaad, N. W., & Stall, R. E. (1988). Initial identification of common genera. In N. W. Schaad (Ed.), Laboratory Guide for identification of plant pathogenic bacteria (2nd ed., pp. 81–84). Phytopathlogy.
- Studholme, J., Wicker, E., Abrare, M., Aspin, A., Bogdanove, A., Broders, K., Dubrow, Z., Grant, M., Jones, B., Karamura, G., Lang, J., Leach, J., Mahuku, G., Nakato, V., Coutinho, J., Smith, J., & Bull, T. (2020). Transfer of Xanthomonas campestris pv. arecae and X. campestris pv. musacearum to X. vasicola (Vauterin) as X. vasicola pv. arecae comb. nov. and X. vasicola pv. musacearum comb. nov. and Description of X. vasicola pv. vasculorum pv. nov. https://doi.org/10.1094/PHYTO-03-19-0098-LE
- Tariku, H., Kassahun, S., Endale, H., & Mengistu, O. (2015). Evaluation of enset clones resistance against enset bacterial wilt disease (Xanthomonas campestris pv. musacearum). Veterinary Science & Technology, 6(1), 3.
- Tesfaye, B. (2008). On Sidama folk identification, naming and classification of cultivated enset (Ensete ventricosum). Genetic Resource and Crop Evaluations, 55(8), 1359–1370. https://doi.org/10.1007/s10722-008-9334-x
- Tripathi, L., Odipio, J., Tripathi, J. N., & Tusiime, G. (2008). A rapid technique for screening banana cultivars for resistance to Xanthomonas wilt. European Journal of Plant Pathology, 121(1), 9–19. https://doi.org/10.1007/s10658-007-9235-4
- Tushemereirwe, W., Kangire, A., Smith, J. J., Ssekiwoko, F., Nakyanzi, M., Kataama, D., Musiitwa, C., & Karyaija, R. (2003). An outbreak of bacterial wilt on banana in Uganda. InfoMusa, 12(2), 6–8.
- Van der Plank, J. E. (1963). Plant diseases: Epidemics and control. Academic Press.
- Wheeler, B. E. (1969). An introduction to plant diseases. Wiley and Sons.
- Winstead, N., & Kelman, A. (1952). Inoculation techniques for evaluating resistance to Pseudomonas solanacearum. Phytopathology, 42(1), 628–634.
- Yemane, T., & Fassil, K. (2006). Diversity and cultural use of Enset (Enset ventricosum (Welw.) Cheesman) in Bonga in situ Conservation Site, Ethiopia. Ethnobotany Research & Applications, 4(1), 147–157. https://doi.org/10.17348/era.4.0.147-158
- Zerihun, Y., Hussein, M., Mulugeta, D., Temesgen, A., & Guy, B. (2013). Enset (Ensete ventricosum) clone selection by farmers and their cultural practices in southern Ethiopia. Genetic Resources and Crop Evolution, 61(3), 1–16.