Abstract
Cassava mosaic disease (CMD), arguably the most important viral disease of cassava, an important food security crop has in recent times, been reported to be associated with satellites in some parts of sub-Saharan Africa. We investigated the status of Cassava mosaic begomovirus and associated satellites in CMD-affected cassava plants in Ghana using polymerase chain reaction. In all, 110 CMD-affected cassava leaf samples collected from cassava fields in Southern Ghana. Africa cassava mosaic virus (ACMV) was detected in 107 (97%) cassava leaf samples, out of which 76 (69%) was in mixed infections with East Africa cassava mosaic virus-Cameroon (EACMV-CM). EACMV (CM) alone was detected in three (3%) cassava leaf samples. Out of the 110 cassava leaf samples affected by cassava mosaic begomoviruses, satIII and satII were detected in 66 (60%) and 47 (43%), respectively. To the best of our knowledge, detection of CMB-associated satellites in Ghana is the first in the country and possibly in West Africa, and has serious epidemiological implications on the management of CMD, thereby requiring further studies and concerted efforts to safeguard cassava production.
PUBLIC INTEREST STATEMENTS
Cassava, known as “agbeli” in the Ewe dialect in West Africa, is a major a food security crop in Ghana and much of sub-Saharan Africa. Cassava is a good source of income and an important crop to the vulnerable and disadvantaged. Apart from its uses for food (many local diets), it has several industrial uses as ethanol, biofuel, cassava flour, cassava chips, starch, and wet cake. Despite the importance of cassava, its production is hindered by cassava mosaic disease (CMD), a disease caused by viruses. This disease reduces the yield of cassava significantly. We reported in Ghana, in addition to cassava mosaic begomoviruses, particles that are much smaller than viruses.These particles are known as satellites, and are reported to make cassava plants more susceptible to the disease. This current finding could influence policy leading to finding cassava planting materials that will produce optimum yield in the future.
1. Introduction
Cassava (Manihot esculenta Crantz) is one of the important crops in sub-Saharan Africa (FAO, Citation2010) and has become a good income source to families, peasant and subsistence farmers all year round. It's a good food security crop in many African countries. Today, cassava has become one of the major staple crops cultivated across sub-Saharan Africa, producing 177 million metric tons (MMT) of the total world production of 284.9 MMT (FAOSTAT, Citation2017).
Ghana is the third highest producer of cassava in Africa, producing 18 MMT (FAOSTAT, Citation2017). Cassava is the most important staple in the country, with per capita daily intake of 642 calories, far exceeding maize and rice with 434 and 217 calories, respectively (FAO, Citation2006).
Despite its enormous importance, cassava cultivation is constrained by cassava mosaic disease (CMD). CMD is arguably described as the most important disease of the crop in Africa (Legg & Fauquet, Citation2004; Sseruwagi et al., Citation2004). CMD is caused by several species of single-stranded DNA viruses in Africa and Asia, usually referred to as Cassava mosaic begomoviruses (CMBs), family, Geminiviridae (Sseruwagi et al., Citation2004), and include African cassava mosaic virus (ACMV), East African cassava mosaic virus (EACMV), South African cassava mosaic virus (SACMV) and Indian cassava mosaic virus (ICMV) (Brown et al., Citation2015). These viruses are transmitted by whiteflies [Bemisia tabaci (Gennadius)] and perpetuated through infected cuttings, which is the usual method of propagating the crop (Otim-Nape et al., Citation1994).
Begomoviruses are often associated with sub-viral agents referred to as alphasatellites and betasatellites. Until recently, most of the single-stranded DNA satellites were reportedly associated with monopartite begomoviruses (Patil & Fauquet, Citation2010), and are about 1.3 kb in size. Alphasatellites encode their own Rep and replicate autonomously (Ndunguru et al., Citation2016). Betasatellites, on the other hand, depend on begomoviruses for their replication, movement and encapsidation (Mubin et al., Citation2020). Betasatellites are diverse set of symptom-enhancing molecules that are associated with monopartite begomoviruses (Briddon et al., Citation2008; Gnanasekaran et al., Citation2019).
During the last two decades, unusual disease symptoms, such as filiform-shaped leaves, seen on CMB-infected cassava in East Africa, led to the discovery and description of satellites from cassava that are different from both alphasatellites and betasatellites (Ndunguru et al., Citation2016, Citation2008). Similar reported occurrence of satellites together with bipartite, cassava mosaic begomoviruses on CMD affected cassava plants in East and Central Africa had also been made (Mwatuni et al., Citation2015; Ndomba, Citation2012) with major implications for epidemiology of CMD. Earlier studies on CMD in Ghana has reported the occurrence of species of Cassava mosaic begomovirus but not satellites (Torkpo et al., Citation2017). The present study therefore seeks to investigate the status of satellites and Cassava mosaic begomoviruses that are associated with cassava in the Ghana, a major cassava producer in sub-Saharan Africa.
2. Materials and methods
2.1. Collection of CMD-affected cassava leaves
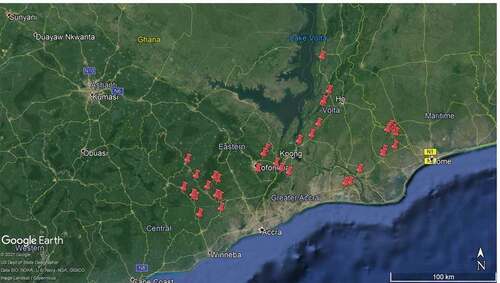
Young leaves from CMD-affected plants were collected in 2018/19 in parts of Southern Ghana, namely: Eastern, Central, Volta, and Greater Accra regions for the study (). At each location, an observation was made of the presence or absence of CMD using modified “Z” fashion (i.e., from two sides of the field and along a diagonal across the field (Otim-Nape et al., Citation1998). Sample collection was not limited to only major cassava growing areas. Young moderately severe to very severe CMD affected leaves were collected into plastic bags, labelled and kept on ice and transported to the Biotechnology Centre of the School of Agriculture in the University of Ghana. DNA extraction and further laboratory analysis were performed on 110 samples collected.
2.2. DNA extraction and polymerase chain reaction (PCR)
DNA was extracted from the leaf samples using the CTAB method (Doyle & Doyle, Citation1987) and quality of isolated DNA assessed by electrophoresis prior to PCR (Saiki et al., Citation1988). PCR was performed using specific primer pairs () designed to detect ACMV, EACMV (CM), satII and satIII. The reaction mixture comprised of 12.5 µL Master Mix (New England Biolab, NEB), 1.0 µL each of forward and reverse primers (10 pM), 8.5 µL nuclease free PCR water (Inqaba Biotech West Africa Ltd) and 2.0 µL DNA to make up a final volume of 25.00 µL. The cycling regime for the amplification was as follows: initial denaturation at 94°C for 3 min, followed by 30 cycles of 94°C for 1 min, 56°C for 1 min 30 sec and 72°C for 1 min, with a final extension of 72°C for 4 min (Mwatuni et al., Citation2015) in the Applied Bio systems Thermal Cycler. Amplicons were separated by electrophoresis on 1.5% agarose gels (which was stained with ethidium bromide) for 100 V for 1 h. Bands were viewed and the images saved using Syngene Gel documentation system.
Table 1. Primers, primer sequences and their respective target viruses used for the Cassava mosaic begomovirus and associated satellites
3. Results and discussion
Even though asymptomatic cassava plants were recorded on some of the locations, leaves of cassava plants showing moderately severe to very severe cassava mosaic symptoms () were the samples collected and analyzed. DNA was successfully obtained from all the samples collected. ACMV, EACMV-CM, satellite DNA II (satII) and satellite DNA III (satIII) were detected from the DNA extracted from the leaf samples in PCR using specific primers
Figure 2. Cassava leaves. (a), Asymptomatic (b), Moderately severe mosaic symptom (c), Severe mosaic symptom (d). Very severe mosaic symptom

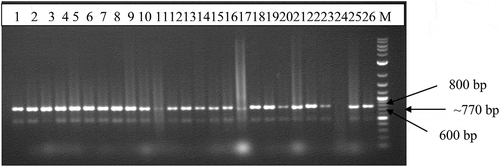
Cassava mosaic begomoviruses were detected in all the cassava leaf samples. The presence or absence of Cassava mosaic begomovirus or DNA satellites in a sample was not dependent on the type of symptom. Out of 110 CMD-affected samples tested, 107 tested positive for ACMV (), and 76 tested positive for EACMV-CM. This represented 97.0% and 69.0% of samples that tested positive for ACMV and EACMV-CM, respectively. Three (3) of the CMD-affected samples were singly infected by only EACMV. This was equivalent to 2.7% of the samples. Thirty-one (31) of the samples were positive for only ACMV.
Figure 3. Image of electrophoresis of total PCR product on 1.5% agarose gel generated using primer pair JSP001/002 that detect ACMV. M-1.0 kb molecular marker; Lanes represent CMD-affected leaf samples collected from the various locations in southern Ghana
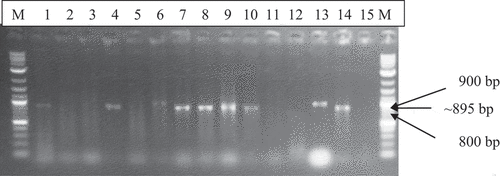
Overall, satII and satIII were detected in 47 and 66 of the CMD-affected cassava leaves, representing 43% and 60% respectively. The expected PCR products of ~895 bp and ~306 bp were respectively obtained with the satII F/R and satIII F/R primers. The 1.0 kb molecular marker (GIBCO Life Technologies), which was correctly used, gave the expected PCR product (). Both satellites were widely distributed in southern Ghana. A gel image of severely CMD-affected cassava leaf samples arranged in the same order is presented for satIII () and satII ().
Figure 4. Agarose gel electrophoresis of DNA satIII specific PCR products of 306 bp from CMD-affected cassava leaf samples total nucleic acid. Lanes 1 to 14 contained separate cassava samples. M-1.0 kb molecular marker; Lane 15- Negative Control
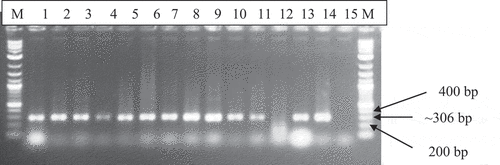
Figure 5. Agarose gel electrophoresis of DNA satII specific PCR products of 895 bp from CMD-affected cassava leaf samples total nucleic acid. Lanes 1 to 14 . Lanes 1 to 14 contained separate cassava samples. M-1.0 kb molecular marker; Lane 15- Negative Control
ACMV was detected in the majority of CMD-affected cassava leaves, and remain the predominant Cassava mosaic begomovirus species in the country, which is consistent with earlier findings in the country (Offei et al., Citation1999; Oteng-Frimpong et al., Citation2012; Torkpo, Citation2009; Torkpo & Offei, Citation2007) in which they reported ACMV as the frequently detected CMB in the country. ACMV was also the predominant CMB species in Nigeria (Alabi et al., Citation2007; Ogbe et al., Citation2006) and Togo (Adjata et al., Citation2009). The Mixed infection of ACMV and EACMV (CM) as revealed in the present study concurs with that of other parts of Africa in which more the one CMB species were reported from CMD-affected cassava plants. (Adjata et al., Citation2009; Alabi et al., Citation2007; Busogoro et al., Citation2008; Ogbe et al., Citation2006; Were et al., Citation2004).
Detection of single infection of cassava by EACMV in the current study agrees with Offei et al. (Citation1999) but not other findings in the country (Torkpo & Offei, Citation2007; Torkpo et al., Citation2017), in which no single EACMV infection was detected in the country.
We report, for the first time, the occurrence of DNA satII and DNA satIII in Ghana, and in West Africa. More samples tested positive for DNA satIII compared to DNA satII, and concured with what occurred in Tanzania (Ndomba, Citation2012). Mwatuni et al. (Citation2015) reported high occurrence of DNA satIII in Kenya though it is less in comparison to what was reported in this present study. Additionally, higher levels of DNA satII reported in the present study contradicts the very low numbers reported by Mwatuni et al. (Citation2015) but not Ndomba Citation2012. The occurrence of high levels of DNA satII and DNA satIII in Ghana presents major challenges to the already serious CMD situation. Sub-viral agents have been reported from CMD-affected plants in Tanzania (Ndunguru et al., Citation2016, Citation2008), and the authors stressed that the sub-viral agents have the ability to overcome CMD2 resistance, and their whitefly transmissibility has major implications for the long-term durability of CMD2 resistance. The epidemiological implications of these sub-viral agents reported in Ghana on the country’s production of cassava cannot, therefore, be over emphasized.
4. Conclusion
ACMV remains the predominant Cassava mosaic begomovirus species and was detected in all the 46 locations in the south of the country. EACMV-CM was detected in 35 locations. Mixed ACMV and EACMV-CM was detected. Three cassava leaf samples were positive for EACMV-CM alone. DNA Satellites, satIII and satII were detected in 25 and 17 locations, respectively. Both satIII and satII were detected in mixed infections with CMBs. No DNA Satellite was detected alone from any cassava leaf sample. Detection of CMBs, satIII and satII was across sample type. This was the first detection of DNA Satellites from CMD-affected cassava leaves in the country. This undoubtedly would have epidemiological implications that could pose challenges to CMD management and have negative effect on cassava yield and food security.
Acknowledgments
The authors are grateful to staff of the Biotechnology Centre of the University of Ghana, Legon for their support during the laboratory work.
Additional information
Funding
Notes on contributors

Stephen Kwame Torkpo
Stephen K. Torkpo holds a PhD (Crop Science) from the University of Ghana, Legon. He is a Research Fellow at the University of Ghana. His research focuses on diagnosis, epidemiology and management of viral and viral-like diseases of crops using conventional and molecular techniques. He was involved with the laboratory work and drafting of the manuscript.
Emmanuel Amponsah
Emmanuel Amponsah is currently a Masters student at the Department of Crop Science of the University of Ghana, Legon. He contributed to the laboratory work and drafting of the manuscript.
Celestina Danso Arhin
Celestina Danso Arhin holds a Bachelor of Science degree in Agriculture (Crop Science). She contributed to the laboratory work and drafting of the manuscript.
Samuel Kwame Offei
Samuel Kwame Offei is a Professor of Plant Virology and Molecular Biology at the University of Ghana. His research covers all aspect of Plant Virology and Molecular Biology. These include cassava mosaic begomoviruses and satellites..
References
- Adjata, K. D., Muller, E., Peterschmitt, M., Traoré, O., & Gumedzoe, Y. M. (2009). Molecular evidence for the association of a strain of Uganda variant of East African Cassava Mosaic Virus to Symptom Severity in Cassava (Manihot esculenta Crantz) Fields in Togo. American Journal of Biochemistry and Biotechnology, 5(4), 196–9. https://doi.org/https://doi.org/10.3844/ajbbsp.2009.196.201
- Alabi, O., Ogbe, F., Bandyopadhyay, R., Dixon, A. G. O., Hughes, J., & Rayapati,N. (2007). The occurrence of African cassava mosaic virus and East African cassava mosaic Cameroon virus in natural hosts other than cassava in Nigeria. Phytopathology, 97 (7):S3-S3. https://apsjournals.apsnet.org/doi/pdf/10.1094/PHYTO.2007.97.7.S1
- Briddon, R. W., Brown, J. K., Moriones, E., Stanley, J., Zerbini, M., Zhou, X., & Fauquet, C. M. (2008). Recommendations for the classification and nomenclature of the DNA-β satellites of begomoviruses. Archives of Virology, 153(4), 763–81. https://doi.org/https://doi.org/10.1007/s00705-007-0013-6
- Brown, J. K., Zerbini, F. M., Navas-Castillo, J., Moriones, E., Ramos-Sobrinho, R., Silva, J. C., Fiallo-Olive, E., Briddon, R. W., Hernández-Zepeda, C., Idris, A., Malathi, V. G., Martin, D. P., Rivera-Bustamante, R., Ueda, S., & Varsani, A. (2015). Revision of Begomovirus taxonomy based on pairwise sequence comparisons. Archives of Virology, 160(6), 1593–619. https://doi.org/https://doi.org/10.1007/s00705-015-2398
- Busogoro, Jean-Pierre ., Masquellier, L., Kummert, J., Dutrecq, O., Lepoivre, P., & Jijakli, M. (2008). Application of a S implified Molecular Protocol to Reveal Mixed Infections with Begomoviruses in Cassava. Journal of Phytopathology, 156(7–8), 452–457. https://doi.org/https://doi.org/10.1111/j.1439-0434.2008.01391.x
- Doyle, J. J., & Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin, 19(1), 11–15.
- FAO. (2006). FAO year book on Ghana.
- FAO. (2010). Food and Agriculture Organization.
- FAOSTAT. (2017). http://faostat.fao.org
- Fondong, V. N., Pita, J. S., Rey, M. E., De Kochko, A., Beachy, R. N., & Fauquet, C. M. (2000). Evidence of the synergism between African cassava mosaic virus and a new double-recombinant geminivirus infecting cassava in Cameroon. Journal of General Virology, 81 (1), 287–297. DOI:https://doi.org/10.1099/0022-1317-81-1-287
- Gnanasekaran, P., KishoreKumar, R., Bhattacharyya, D., Vinoth Kumar, R., & Chakraborty, S. (2019). Multifaceted role of geminivirus associated betasatellite in pathogenesis. Molecular Plant Pathology, 20(7), 1019–1033. https://doi.org/https://doi.org/10.1111/mpp.12800
- Legg, J. P., & Fauquet, C. M. (2004). Cassava mosaic geminiviruses in Africa. Plant Molecular Biology, 56(4), 585–99. https://doi.org/https://doi.org/10.1007/s11103-004-1651-7
- Mubin, M., Ijaz, S., Nahid, N., Hassan, M., Younus, A., Qazi, J., & Nawaz-Ul-Rehman, M. S. (2020). Journey of begomovirus betasatellite molecules: From satellites to indispensable partners. Virus Genes, 56(1), 16–26. https://doi.org/https://doi.org/10.1007/s11262-019-01716-5.
- Mwatuni, F., Ateka, E., Karanja, L., Mwaura, S., & Obare, I. (2015). Distribution of cassava mosaic geminiviruses and their associated DNA satellites in Kenya. American Journal of Experimental Agriculture, 9(3), 1–12. https://doi.org/https://doi.org/10.9734/AJEA/2015/18473
- Ndomba, A. O. (2012). ‘Influence of satellite DNA molecules severity of cassava begomoviruses and the breakdown of resistance to cassava mosaic disease in Tanzania [PhD Thesis]. University of the Witwatersrand.
- Ndunguru, J., De León, L., Doyle, C. D., Sseruwagi, P., Plata, G., Legg, J. P., Thompson, G., Tohme, J., Aveling, T., Ascencio-Ibáñez, J. T., & Hanley-Bowdoin, L. (2016). Two novel DNAs that enhance symptoms and overcome CMD2 resistance to cassava mosaic disease. Journal of Virology, 90(8), 4160–4173. https://doi.org/https://doi.org/10.1128/JVI.02834-15
- Ndunguru, J., Fofana, B., Legg, J. P., Challappan, P., Taylor, N., Aveling, T., Thomson, G., & Fauquet, C. M. (2008, July 21- 25). Two novel satellite DNAs associated with bipartite cassava mosaic begomoviruses enhancing symptoms and capable of breaking high virus resistance in 95 cassava landraces. Proceeding of the First Scientific Meeting of the Global Cassava Partnership (GCP-I), Ghent, Belgium.
- Offei, S. K., Owuna-Kwakye, M., & Thottappilly, G. (1999). First report of East African cassava mosaic begomovirus in Ghana. Plant Disease, 83(9), 877. https://doi.org/https://doi.org/10.1094/PDIS.1999.83.9.877C
- Ogbe, F. O., Dixon, A., Hughes, Jd’A, Alabi, O. J., & Okechukwu, R. (2006). Status of cassava begomoviruses and their new natural hosts in Nigeria. Plant Disease, 90(5), 548–553. https://doi.org/https://doi.org/10.1094/PD-90-0548
- Oteng-Frimpong, R., Levy, Y., Torkpo, S. K., Danquah, E. Y., Offei, S. K., & Gafni, Y. (2012). Complete genome sequencing of two causative viruses of cassava mosaic disease in Ghana. Acta virologica, 56(4), 305–314. https://doi.org/https://doi.org/10.4149/av_2012_04_305
- Otim-Nape, G. W., Shaw, M. W., & Thresh, J. M. (1994). The effects of African cassava mosaic geminivirus on the growth and yield of cassava in Uganda. Tropical Science, 34 (Special Issue), 43–54.
- Otim-Nape, G. W., Thresh, J. M., & Shaw, M. W. (1998). The incidence and severity of cassava mosaic virus disease in Uganda: 1990-92. Tropical Science, 38 (1), 25–37.
- Patil, B. L., & Fauquet, C. M. (2010). Differential interaction between cassava mosaic geminiviruses and geminivirus satellites. Journal of General Virology, 91(7), 1871–82. https://doi.org/https://doi.org/10.1099/vir.0.019513–0
- Saiki, R. K., Gelfand, D. H., Stoffel, S., Scharf, S. J., Higuchi, R., Horn, G. T., Mullis, K. B., & Erlich, H. A. (1988). Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science, 239(4839), 487–-91. https://doi.org/https://doi.org/10.1126/science.239.4839.487
- Sseruwagi, P., Sserubombwe, W. S., Legg, J. P., Ndunguru, J., & Thresh, J. M. (2004). Methods of surveying the incidence and severity of cassava mosaic disease and whitefly vector populations in Africa: A review. Virus Research, 100(1), 129–142. https://doi.org/https://doi.org/10.1016/j.virusres.2003.12.021
- Torkpo, S. K. (2009). Analysis of cassava mosaic begomoviruses associated with cassava mosaic disease in Ghana [PhD Thesis]. University of Ghana.
- Torkpo, S.K., Offei, K., Danquah, E.Y., & Gafni, Y. (2017). Status of cassava mosaic begomoviruses in farmers’ fields in Ghana. AIMS Agriculture and Food, 2(3), 279–289. https://doi.org/https://doi.org/10.3934/agrfood.2017.3.279
- Torkpo, S. K., & Offei, S. K. (2007). Status of cassava mosaic disease in farmers’ fields. Book of abstracts of the 25th biennial Conference of the Ghana Science Association held at Bunso and Tafo in the Eastern Region of Ghana between August 5-10, 2007.
- Were, H., Winter, S., & Maiß, E. (2004). Variations and taxonomic status of begomoviruses causing severe epidemics of cassava mosaic disease in Kenya, Uganda, and Democratic Republic of the Congo. Journal of General Plant Pathology, 70(4), 243–248. https://doi.org/https://doi.org/10.1007/s10327-003-0119-y