Abstract
Background: The recessive rough fur mutation (ruf)―named for the unkempt, greasy appearance of the hair coat in homozygotes―has previously been mapped on mouse Chromosome 9. However, the assignment of ruf to a particular gene is needed to facilitate a complete molecular analysis of the mutant phenotype. Results: To establish a more refined location for ruf (as a basis for positional cloning) DNA isolated from a large backcross family was typed for microsatellite and single-nucleotide markers on Chromosome 9. This analysis restricted the location of ruf between sites that flank only four genes known to be expressed in skin, one of which―Mpzl3, for myelin protein zero-like 3―generates a similar hair phenotype in mice homozygous for engineered and spontaneous null-alleles. A cross between ruf mutants and mice heterozygous for the Mpzl3rc mutation (which controls a recessive phenotype called rough coat) produced offspring that displayed matted, damp-looking fur, indicating that ruf is a mutant allele of Mpzl3. However, sequence analysis of the Mpzl3 promoter, exons and splice junctions revealed no mutant-specific DNA defect. Conclusion: The results presented indicate that ruf is a mutant allele of Mpzl3. With a genetic assignment in hand, the rough fur variant can now be more fully characterized to advance our understanding of Mpzl3’s role in normal skin development and function, hepatic triglyceride synthesis, weight regulation, energy and glucose homeostasis; and to model-related human disorders.
Public Interest Statement
The rough fur mouse variant has been known for more than 30 years, but its detailed analysis has been hampered by a lack of molecular probes. Here, we used a positional approach to identify a small number of gene candidates, and complementation testing to assign ruf to the Mpzl3 gene. This work places the rough fur mutant among a series of other known Mpzl3 variants that, together, comprise a living resource that promises to advance a complete molecular analysis of Mpzl3’s complex role in mammalian physiology, and offers a model system for the study of orthologous human conditions that may be characterized by alopecia, over-active sebaceous glands, or altered glucose homeostasis and weight regulation.
Competing Interests
The authors declare no competing interest.
1. Introduction
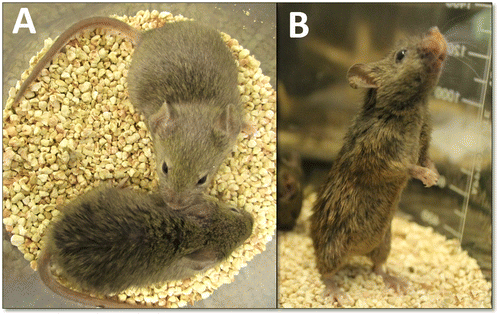
The recessive rough fur mutation in mice (abbreviated ruf) generates a hair coat in homozygotes that looks unkempt and greasy by weaning age (see Figure ) and persists thereafter. This mutation arose spontaneously in the C3H/HeJ inbred strain at The Jackson Laboratory (Bar Harbor, ME, USA) in 1985, and its initial description (including frequently broken hairs and mild hyperkeratosis) and linkage assignment to Chromosome (Chr) 9 were published by Sweet et al. (Citation1990). Light and electron microscopic evaluation of the pelage skin from mutants by Park et al. (Citation2001) showed large lipid deposits on the surface of the epidermis and in abnormally large sebaceous glands, suggesting that the rough fur mutant may be an animal model for a human disorder characterized by hyperlipidogenesis, but no particular disorder was named.
Figure 1. The rough fur phenotype. (A) A 60-day-old mutant (ruf/ruf) female is shown (bottom) with a phenotypically normal (heterozygous) sister (top). (B) The same mutant female is shown, standing upright.
To facilitate assignment of the mutant rough fur phenotype to a specific genetic cause―which would provide molecular probes for the further analysis of this interesting phenotype and help to identify an orthologous human disorder―here we have fine-mapped the ruf mutation with respect to various microsatellite and single-nucleotide polymorphisms on mouse Chr 9. This analysis identified a small set of co-localizing, skin-expressed candidate genes, and ruf has been found (by complementation testing) to be an allele of one of these, Mpzl3 (for myelin protein zero-like 3).
2. Materials and methods
Animals were housed and fed according to Federal guidelines, and the Institutional Animal Care and Use Committee at Central Connecticut State University (CCSU) approved of all procedures involving mice (Animal Protocol Applications 151 and 153). Mice from the standard inbred strains C57BL/6 J (Jax Stock Number 664) and C3H/HeJ (Jax Stock Number 659), inbred C3H/HeJ-ruf/J mice (Jax Stock Number 1544, segregating for ruf), and C57BL/6 J-Mpzl3rc/J mice (Jax Stock Number 540, segregating for rough coat, Mpzl3rc) were obtained from The Jackson Laboratory (Bar Harbor, ME, USA). Both mutant strains were derived from cryopreserved stocks that were imported and then maintained at CCSU in testcrosses. Both mutants were reliably identified by the presentation of matted, greasy hair at weaning age.
Genomic DNA was isolated from 2 mm tail-tip biopsies taken from two- to three-week-old mice using Nucleospin® Tissue kits distributed by Clontech Laboratories, Inc. (Mountain View, CA, USA), as directed. The polymerase chain reaction (PCR) was performed in 13 μl reactions using the Titanium PCR kit from Clontech Laboratories, as directed. Oligonucleotide primers for PCR were designed and synthesized by Integrated DNA Technologies, Inc. (Coralville, IA, USA), based on sequence information available online (Ensembl Mouse Genome Server, Citation2017; Mouse Genome Database [MGD], Citation2017). To score PCR product sizes for seven different microsatellite markers on Chr 9 (Dietrich et al., Citation1996), reactions plus 2 μl loading buffer were electrophoresed through 3.5% NuSieve® agarose (Lonza, Rockland, ME, USA) gels. Gels were stained with ethidium bromide and photographed under ultraviolet light (see Supplementary Figure S1 for typical results). In addition, four DNA markers based on single nucleotide polymorphisms (SNPs) previously reported to differ between strains C3H/He and C57BL/6 (MGD, Citation2017) were also scored. These markers (herein designated SNPA-D) are described in detail in Supplementary Tables S1 and S2 (and typical results are shown in Supplementary Figure S2). For DNA sequence analysis, about 1.5 μg of individual PCR amplimers were purified and concentrated into a 30 μl volume using QIAquick® PCR Purification kits (Qiagen Sciences, Germantown, MD, USA) prior to primer-extension sequence analysis performed by SeqWright DNA Technology Services (Houston, TX, USA) or by the Keck Foundation Resource Laboratory at Yale University (New Haven, CT, USA).
The Mpzl3rc mutation is a G to A transition at position 9:45066394 that is predicted by Cao et al. (Citation2007) to cause the substitution of a conserved, basic arginine at position 100 with a polar glutamine. This point mutation also destroys an AlwI target site. Carriers of the recessive Mpzl3rc allele were thus identified by endonuclease treatment of a 370 bp PCR product directed by forward (5′ GACAGCCAAGGGAAGAGAAG 3′) and reverse (5′GGAGTCACAGCATGAGAAGAG 3′) primers that flank Exon 3, which contains the rc mutation (see Supplementary Figure S3).
Total RNA was isolated from skin samples taken from 1-month-old mutant and control mice using the Nucleospin® RNA Midi kit by Macherey-Nagel (Bethlehem, PA, USA). From these total RNA samples, poly-A+ mRNA was purified using the NucleoTrap® mRNA kit (also by Macherey-Nagel), and cDNA was generated using the SMARTer® RACE 5′/3′ kit (Clontech Laboratories, Inc.). To detect Mpzl3-specific sequences, primer pairs that flanked exon-junction boundaries [Exon 2 (5′-GTGCGCGTATCGTCCTCTCCTTG-3′) and Exon 4 (5′-TCTTATAGCCAGACCTGCTCCTCTTCTG-3′), or Exon 4 (5′-GCTGTCTTCTGTGGCCCTTCTCTC-3′) and Exon 6 (5′-GTCATGAGTCCCTTCTGCGAGTTTCC-3′)] were used to direct standard PCR amplifications of these cDNAs. To detect mouse β actin (Actb) transcripts, a primer pair within Exon 4 (5′-CCCAGCCATGACGTAGCCATCCA-3′ and 5′-GAAGCTGTAGCCACGCTCGGTCAG-3′) was used. The resulting products were visualized in 3% NuSieve® agarose gels. For primer-extension sequencing, these products were purified and concentrated (as described above) and shipped to SeqWright DNA Technology Services or the Keck Foundation Resource Laboratory at Yale University.
3. Results
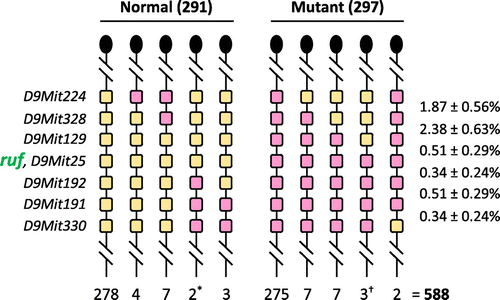
The linkage of ruf on mouse Chr 9, about 3.5% recombination centromeric to Apoa1, was reported by Sweet et al. (Citation1990), but its location has not been defined with respect to other molecular markers. To further refine ruf’s chromosomal location, we produced a 588-member backcross (N2) family by breeding (C57BL/6 J x C3H/HeJ-ruf/J)F1 females with C3H/HeJ-ruf/J mutant males. These N2 mice were typed for their hair phenotype, and DNA isolated from each mouse was characterized for various PCR-scorable, microsatellite markers (Dietrich et al., Citation1996; and see Figure S1) that lie near and centromeric to Apoa1. This haplotype analysis (summarized in Figure ) positioned ruf between markers D9Mit129 and D9Mit192, very near marker D9Mit25 (C3H/HeJ- or C57BL/6 J-derived alleles of which were never meiotically separated from ruf or ruf+ alleles, respectively, in this backcross panel).
Figure 2. Segregation of alleles of ruf and 7 dimorphic microsatellite markers among 588 progeny from an intraspecific mouse backcross. Heterozygous F1 mice (C3H/HeJ-ruf/J x C57BL/6 J) were backcrossed to homozygous C3H/HeJ-ruf/J mutants. The resulting progeny were scored for their rough fur phenotype, and a DNA sample from each mouse was typed for the microsatellite markers shown at the left. Only the Chr 9 haplotype inherited from the F1 parent is shown (with a knob at the top of the haplotype representing the centromere), and the number of mice inheriting that haplotype is shown below it. Very similar numbers of heterozygous and mutant progeny (as expected for a testcross, p > 0.8) suggest that the mutant phenotype is both fully penetrant and fully viable. The two phenotypically normal recombinants marked with an asterisk show that the ruf locus must lie centromeric to D9Mit192. The three mutant recombinants marked with a dagger show that ruf must lie telomeric to D9Mit129. Genetic distances in percentage recombination are shown to the right (± 1 Standard Error).
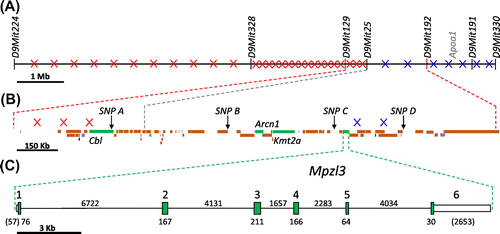
To more precisely locate ruf in this interval, the five mice recombinant between D9Mit129 and D9Mit192 (see Figure (A)) were typed for four single-nucleotide polymorphisms (SNPs) that lie within that interval (and are described in detail in Supplementary Tables S1 and S2, with typical results displayed in Supplementary Figure S2). This analysis, summarized in Figure (B), positioned the three crossovers that fell centromeric to ruf between D9Mit129 and SNP A; and the two crossovers that fell telomeric to ruf were located between SNP C and D. These results show that the ruf mutation must lie between D9Mit129 and SNP D, very near SNP A, B, and C, which (like D9Mit25) were never separated from ruf in this backcross panel. This 1.4-Mb span contains 51 genes or processed transcripts, only four of which (Cbl, for Casitas B-lineage lymphoma; Arcn1, for archain 1; Kmt2a, for lysine (K)-specific methyltransferase 2A; and Mpzl3, for myelin protein zero-like 3) are known to be expressed in skin (MGD, Citation2017)(see Figure (B) and (C)). Each of these candidate genes has been knocked out in mice (Cbl, Thien et al., Citation2003; Arcn1, Xu et al., Citation2010; Kmt2a, McMahon et al., Citation2007), but only mutants homozygous for Mplz3 null alleles (Czyzyk et al., Citation2013; Leiva et al., Citation2014) presented hair and skin anomalies that are similar to the rough fur phenotype.
Figure 3. Physical maps of the ruf region on mouse Chr 9. (A) The relative positions of the seven microsatellite markers typed within the backcross panel are shown (with a 1 Mb scale bar). The crossovers found in each marker-defined interval are shown as a red “x” for crossovers that fell centromeric of ruf, and as a blue “x” for crossovers that fell telomeric to ruf. As described in Figure , the ruf locus must be located between markers D9Mit129 and D9Mit192. (D9Mit25 was never meiotically separated from ruf in this backcross panel.) (B) The relative positions of the four SNP markers typed within the backcross panel are shown for the 1.7 Mb region that is flanked by D9Mit129 and D9Mit192 (with a 0.15 Mb scale bar). Three crossovers that fell between D9Mit129 and SNP A, and cenomeric to ruf, are depicted by red x’s. Two crossovers that fell between SNP C and SNP D, and telomeric to ruf, are depicted by blue x’s. (SNP A, B, and C never separated from ruf in this backcross panel.) These data limit the location of ruf to a 1.4 Mb span between D9Mit129 and SNP D, where 49 protein coding genes (orange and green boxes), 2 processed transcripts (blue boxes), 11 RNA genes, (purple boxes), and 3 pseudogenes (grey boxes) are located. Only 4 of these protein-coding genes (green boxes) are known to be expressed in skin: Cbl, Arcn1, Kmt2, and Mpzl3. (C) The Mpzl3 gene is expanded to show the 6 exons it comprises (with a 3 kb scale bar). Tall green boxes represent coding regions and shorter white boxes indicate the 5′ and 3′ untranslated regions. The number below each coding segment or above each intron is its length in base pairs; the base-pair lengths of noncoding segments are in parentheses. All of the Mpzl3 exons and more than 3,400 bp upstream of the + 1 site were sequenced, but no mutant-specific changes were found.
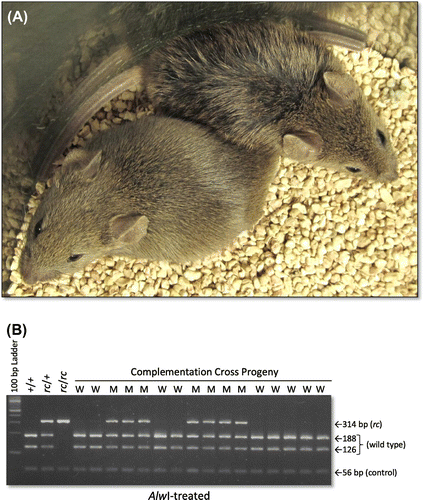
To determine if ruf might be an allele of Mpzl3, we crossed ruf/ruf mutants with mice heterozygous for the spontaneous rough coat mutation (Dickie, Citation1966; Ruvinsky et al., Citation2002), which has been shown to be an allele of Mpzl3 by Cao et al. (Citation2007) and is now designated Mpzl3rc. Because the progeny that inherited both ruf and a recessive Mpzl3rc allele showed matted, greasy hair (while siblings that inherited ruf and Mpzl3+ did not)(see Figure ), we conclude that these recessive mutations “fail to complement”, indicating that ruf is a variant allele of Mpzl3.
Figure 4. The recessive ruf and Mpzl3rc mutations fail to complement in doubly heterozygous mice. A cross between Mpzl3+/Mpzl3rc heterozygotes and ruf/ruf mutants yielded 17 phenotypically normal and 14 progeny with matted hair, suggesting that ruf is a mutant allele of Mpzl3. (A) Representative, 30-day-old progeny from this cross are shown: the mouse on the lower left is phenotypically normal, while the mouse toward the upper right displays damp-looking, matted fur. (B) The Mpzl3rc status of all complementation-test progeny was verified by a DNA test that reveals the rc-specific missense mutation (see Supplementary Figure S3). Lanes that used template DNA from mice with a wild-type phenotype are marked “W”, and those using template DNA from mice with a mutant phenotype are marked “M”. All 31 complementation-cross progeny were genotyped (typical results are shown), and all phenotypic mutants were proven to carry the defective Mpzl3rc allele and one copy of the ruf allele. All phenotypically normal mice carried one copy of ruf, but no copy of Mpzl3rc.
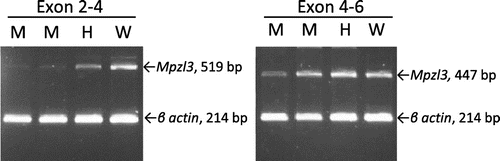
Seeking a ruf-specific defect, we sequenced all 6 exons of the Mpzl3 gene (including both coding and untranslated regions―see Figure (C)), at least 20 base pairs of all intronic splice junctions, and more than 3,400 bp from the promoter region in genomic DNA from ruf/ruf mutants and from control C57BL/6 J and C3H/HeJ mice, but found no sequence differences among any of these templates (data not shown). Next, we isolated poly-A+ mRNA from mutant (ruf/ruf) and control (C3H/HeJ and +/ruf heterozygous) skin, and amplified sequences between Exons 2 and 6 of Mpzl3 (see Figure ). Mutant and control product lengths were the same, and sequence analysis of these amplimers (data not shown) verified the normal splicing of Mpzl3, Exons 2–6, in mutant (and control) skin.
Figure 5. Analysis of Mpzl3 splice junctions in control and mutant (ruf/ruf) cDNA. Poly-A+ RNA from skin was reverse transcribed into cDNA and amplified using forward and reverse primers that anneal with Exon 2 and Exon 4 (left panel), or Exon 4 and Exon 6 (right panel) of Mpzl3. Primers that anneal within Exon 4 of the mouse β actin (Actb) gene were used together with Mzpl3 primers to provide an internal loading control. Templates from ruf/ruf mutants (labeled M above), a ruf/+ heterozygote (labeled H), and a wild type +/+ control (labeled W) all yielded amplimers of the size and sequence expected for normal splicing of Exons 2 through 6. While the intensity of Mpzl3-specific products amplified from mutant templates may appear less abundant than products amplified from control skin samples, it should be noted that the RT-PCR method used here was not quantitative.
Random retroviral insertions have been estimated to be the cause of 10–15% of all spontaneous mutations in mice (Maksakova et al., Citation2006), and more than half of these have occurred on the C3H/He genetic background—where the ruf variant originated. On that basis, we set out to test non-coding regions in and around Mpzl3 for the presence of a large DNA insertion (such as an endogenous retroviral element, ERV) that might be specifically associated with the ruf mutation. To do this, we designed primer pairs for PCR that would generate amplimers of about 800–1,200 bp using wild type C3H/HeJ genomic DNA templates. A primer pair that gives the expected amplimer size in reactions including wild type C3H/HeJ template DNA, but fails to amplify a product from C3H/HeJ-ruf/ruf templates might flank a mutation-specific ERV. We have recently used this “scanning” approach to identify the novel insertion of an ERV element of the IAP family, sub type I∆1, into the Gata3 gene in mice (Connor & King, Citation2016) that is specific to (and the likely cause of) the juvenile alopecia mutation (abbreviated jal) which also occurred spontaneously on the C3H background, and which, like ruf, showed no DNA defect in coding regions (Ramirez et al., Citation2013). However, our scan of Mpzl3 revealed no large, ruf-specific insertion in any of the introns, in more than 4,900 bp downstream of the polyadenylation signal, nor in more than 5,000 bp upstream of the +1 site.
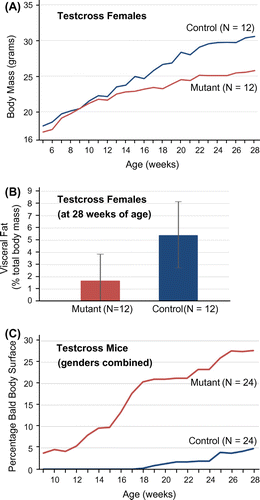
Characterization of other spontaneous (Mpzl3rc, Hayashi et al., Citation2004; Cao et al., Citation2007) or engineered, loss-of-function alleles (Mpzl3tm1(KOMP)Mbp, Czyzyk et al., Citation2013; Mpzl3tm1Tacz, Leiva et al., Citation2014) has revealed a complex array of physiological roles for Mpzl3 that includes regulation of body mass, accumulation of adipose tissue, and progressive alopecia. We therefore measured these attributes in a set of 48 N2 mice (half male, half female; half mutant and half control heterozygotes) from the backcross described above, over the course of 28 weeks. While mutant (ruf/ruf) and control (+/ruf) males gained mass at the same rate, on average (data not shown), mutant females weighed, on average, less than wild-type female controls at all time points, although this difference was not statistically significant (Figure (A)). For example, in week 28, the average mass was 30.6 ± 5.2 g for control females and 25.7 ± 4.6 g for mutant females (± 1 standard deviation). At week 28, these animals were euthanized, abdominal fat was dissected away from other visceral organs, weighed, and recorded as the percentage of total body mass. Again, mutants and controls showed no discernible difference among males, on average (data not shown). By contrast, female mutants showed more than threefold less adipose tissue at 28 weeks of age than heterozygous controls, although this difference was not statistically significant (Figure (B)). Each week during this 28-week study, the percentage of furless body surface (to the nearest 5%) was also estimated. No control mice showed any hair loss prior to week 18, and, by the end of the study, only a few (5/24) showed a modest degree of alopecia (affecting less than 50% of the body surface). By contrast, alopecia appeared in the mutant cohort as early as week 7, and, by the end of the study, most mutants (20/24) showed generally larger bald patches, which affected from 5 to 90% of the body surface (see Figure (C)).
Figure 6. Adiposity and progressive hair loss among N2 mice segregating for rough fur. Backcross mice (24 ruf/ruf and 24 +/ruf) from the mapping panel described in Figure were assessed weekly for body mass (in grams) and for the degree of baldness (estimated as the percentage of hairless body surface). (A) While heterozygous control and mutant males showed no difference in body mass at any point in the 28 week study (data not shown), control females (blue line) showed a markedly (but not significantly) higher mass versus mutant females (red line) at all time points. (B) At the end of the survey, visceral fat was dissected from all N2 subjects, weighed, and expressed as percentage of total body mass. While males showed no difference in fat stores between wild and mutant types (data not shown), control females displayed, on average, three times more visceral fat (5.39 ± 2.76%) than mutants (1.68 ± 2.18%), but this difference was not statistically significant. (C) The estimated size of bald patches (expressed as the percent of the body surface) was averaged among heterozygous control (blue) and mutant (red) N2 mice. Mutants (males and females combined) show a steady, progressive increase in baldness that begins earlier and is more extensive than the hair loss seen among non-mutant control subjects (which was similar to the incidental alopecia typically displayed by C3H/HeJ mice).
4. Discussion
We have taken a positional-candidate approach to assign the classic rough fur mutation in mice to the Mpzl3 gene. Although we could find no corresponding DNA defect in the Mpzl3 gene in rough fur mice, the failure of the recessive rc allele of Mpzl3 to complement the ruf defect in compound heterozygotes confirms allelism. We therefore recommend that the rough fur mutation be formally renamed “myelin protein zero-like 3; rough fur”, with the abbreviation Mpzl3ruf.
We are not the first team to perform a DNA sequence analysis of the Mpzl3 gene in rough fur mutants. Cao et al. (Citation2007) reported no sequence variation in Mpzl3 coding regions in a ruf/ruf DNA sample compared to one from the parental C3H/HeJ strain. Here, we verified that result, but, with reconstituted ruf/ruf mutants that Cao et al. lacked, we were able to test directly for non-complementation and therefore draw a different conclusion (that ruf is an allele of Mpzl3) than Cao and co-workers did in 2007. While our sequence analysis extended beyond the coding regions, more work is needed to determine the precise nature of the Mpzl3ruf defect. We speculate that this defect might, in some way, impact the expression, processing, or stability of the Mpzl3ruf transcript, although we find that the mutant transcript appears to be normally spliced. Quantitative and qualitative evaluation of Mpzl3 transcripts and protein in the epidermis of mutant versus wild-type C3H mice should help to refine this array of possibilities. It is notable that, while we sequenced over 3,400 bp upstream of Mpzl3ruf (and found no defect), others have suggested that the regulatory region for Mpzl3 may extend 12 kb upstream of the transcriptional start site (Boxer, Barajas, Tao, Zhang, & Khavari, Citation2014; Sen et al., Citation2012).
In any case, the spontaneous ruf allele now joins Mpzl3rc and at least two targeted null alleles of Mpzl3 (Czyzyk et al., Citation2013; Leiva et al., Citation2014), providing a collection of phenotypically similar, but mutationally distinct defects. Side-by-side comparison of the phenotypes controlled by each different allele (on similar genetic backgrounds), might reveal important information relating gene structure to function. For example, the spontaneous ruf allele may not show the same pleiotropic effect on adipose accumulation (at least in males) as reported for the targeted Mpzl3 alleles (Czyzyk et al., Citation2013; Leiva et al., Citation2014; Tang et al., Citation2010), but diet and background would need to be controlled and similar assays performed to verify this impression.
The study of spontaneous and targeted mouse models has emphasized Mpzl3’s essential role in skin (where it impacts dermal adipose, hair follicle and sebaceous gland development), hepatic triglyceride synthesis, main adipose deposition, energy expenditure and glycemic control (see Cao et al., Citation2007; Czyzyk et al., Citation2013; Hayashi et al., Citation2004; and Leiva et al., Citation2014). Consistent with these phenotypes, Mpzl3 has recently been found to encode an adhesion protein that is highly induced in epidermal differentiation, and is primarily located in the mitochondria (Bhaduri et al., Citation2015). MPZL3 has been found to interact with numerous mitochondrial proteins including ferrodoxin reductase (FDXR), an enzyme that, together with MPZL3, is necessary for induction of reactive oxygen species (ROS) required for epidermal differentiation (Bhaduri et al., Citation2015). While no humans with MPZL3 deficiencies have yet been reported, based on the described series of spontaneous-mutant and knock-out mouse models it may be predicted that such conditions might present with altered metabolic physiology (such as decreased adiposity and increased food intake in response to metabolic demand), sebaceous gland hypertrophy (as in acne vulgaris) and hair loss (as in alopecia areata). Notably, one inherited skin condition in humans, seborrhea-like dermatitis with psoriasiform elements (OMIM #610227, Online Mendelian Inheritance in Man [OMIM], Citation2017), results from mutation of ZNF750 (Birnbaum et al., Citation2006), which encodes a transcription factor that binds to the MPZL3 promoter to activate its transcription (Bhaduri et al., Citation2015). The readily-available Mpzl3 mutants can clearly serve as useful models to advance our understanding of seborrhoeic dermatitis in general and of ZNF750 dysfunction in particular (see Wikramanayake et al., Citation2016), even in the absence of a Zfp750 knock-out mouse.
Other tissues—including the brain; the tongue, esophagus, stomach, colon and gall bladder; the trachea and lungs; lymph nodes and white blood cells; the thyroid; and the prostate—also display high levels of myelin protein zero-like 3 expression in both mouse and man (Fagerberg et al., Citation2014; Mouse ENCODE Consortium, Citation2014), and the role of Mpzl3 in the development, maintenance and function of these tissues certainly deserves further investigation. We anticipate that mouse variants with distinct defects in Mpzl3―a set which now includes Mpzl3ruf―will help to facilitate such studies.
5. Conclusions
The recessive ruf mutation in mice maps within a 1.4 Mb interval on mouse Chr 9, and has been shown by complementation testing to be a mutant allele of the Mpzl3 gene. While further study will be needed to pinpoint the molecular defect that is the basis of this mutant phenotype, this assignment places ruf among a series of distinct mutant alleles of Mpzl3. This set of mouse variants will surely be valuable as a model system for the in-depth, molecular analysis of the various, pleiotropic functions controlled by Mpzl3, and for the study of some inherited human disorders where those functions are impaired.
Author’s contributions
TRK conceived of the study, carried out all procedures involving mice, and drafted the manuscript. Student coauthors led the various aspects of the study (as listed below), including experimental design, data acquisition and interpretation. Specifically: KMP and MAR performed the genetic mapping analysis; LAR performed the complementation analysis; TVN, MMR, SE, and JJS sequenced the Mpzl3ruf promoter, exons and splice junctions; MEC and MMR scanned Mpzl3ruf for the presence of large insertion elements; TVN conducted the analysis of Mpzl3ruf mRNA; AK performed the time-course analysis of body mass, adipose accumulation and progressive hair loss. All authors read, edited, and approved the final manuscript.
Funding
This work was supported by a small research grant from the Connecticut State Colleges and Universities System.
Supplemental data
Supplemental data for this article can be accessed here https://doi.org/10.1080/23312025.2017.1370058.
| Abbreviations | ||
| Chr | = | chromosome |
| CCSU | = | Central Connecticut State University |
| PCR | = | polymerase chain reaction |
| RT-PCR | = | reverse transcriptase PCR |
| SNP | = | single nucleotide polymorphism |
| ERV | = | endogenous retrovirus |
| IAP | = | intracisternal A particle |
| ROS | = | reactive oxygen species |
OABI_1370058_Supplementary_Material.pdf
Download PDF (962.7 KB)Acknowledgements
The authors thank undergraduate Erin Casey and high school interns Renukanandan Tumu, Brandy Robinson, Tasheika Telfer and Tamara Rodriguez for help with marker typing; and Mary Mantzaris for excellent animal care.
Additional information
Notes on contributors
Thomas R. King
The research group led by Thomas R. King at Central Connecticut State University (New Britain, CT, USA) aims to identify the genetic basis of certain spontaneous hair variants in mice and rats. Identification of such causative gene defects promises to make new probes available for the molecular analysis of the mammalian integument, and to facilitate the identification of human conditions that these animal mutants might model. Mohammed A. Rabah, Tu V. Nguyen, Audra C. Kohm, Morgan M. Ricci, Jachius J. Stewart, and Sidney Eragene were undergraduates when they contributed to this project; Kenneth M. Palanza, Legairre A. Radden II, and Malcolm E. Connor were graduate students in the Department of Biomolecular Sciences.
References
- Bhaduri, A., Ungewickell, A., Boxer, L. D., Lopez-Pajares, V., Zarnegar, B. J., & Khavari, P. A. (2015). Network analysis identifies mitochondrial regulation of epidermal differentiation by MPZL3 and FDXR. Developmental Cell, 35, 444–457.10.1016/j.devcel.2015.10.023
- Birnbaum, R. Y., Zvulunov, A., Hallel-Halevy, D., Cagnano, E., Finer, G., Ofir, R., … Birk, O. S. (2006). Seborrhea-like dermatitis with psoriasiform elements caused by a mutation in ZNF750, encoding a putative C2H2 zinc finger protein. Nature Genetics, 38, 749–751.10.1038/ng1813
- Boxer, L. D., Barajas, B., Tao, S., Zhang, J., & Khavari, P. A. (2014). ZNF750 interacts with KLF4 and RCOR1, KDM1A, and CTBP1/2 chromatin regulators to repress epidermal progenitor genes and induce differentiation genes. Genes & Development, 28, 2013–2026.10.1101/gad.246579.114
- Cao, T., Racz, P., Szauter, K. M., Groma, G., Nakamatsu, G. Y., Fogelgren, B., … Csiszar, K. (2007). Mutation in Mpzl3, a novel (corrected) gene encoding a predicted (corrected) adhesion protein, in the rough coat (rc) mice with severe skin and hair abnormalities. Journal of Investigative Dermatology, 127, 1375–1386.10.1038/sj.jid.5700706
- Connor, M. E., & King, T. R. (2016). The spontaneous juvenile alopecia (jal) mutation in mice is associated with the insertion of an IAP element in the Gata3 gene. Cogent Biology, 2, 1264691.
- Czyzyk, T. A., Andrews, J. L., Coskun, T., Wade, M. R., Hawkins, E. D., Lockwood, J. F., … Statnick, M. A. (2013). Genetic ablation of myelin protein zero-like 3 in mice increases energy expenditure, improves glycemic control, and reduces hepatic lipid synthesis. American Journal of Physiology - Endcrinology and Metabolism, 305, E282–E292.10.1152/ajpendo.00228.2013
- Dickie, M. M. (1966). Rough coat. Mouse News Letter, 34, 30.
- Dietrich, W. F., Miller, J., Steen, R., Merchant, M. A., Damron-Boles, D., Husain, Z., … O’Connor, T. J. (1996). A comprehensive genetic map of the mouse genome. Nature, 380, 149–152.10.1038/380149a0
- Ensembl Mouse Genome Server. (2017). Mouse genome sequencing consortium: The European Bioinformatics Institute (EBI) and Welcome Trust Sanger Institute (WTSI). Release 89. Retrieved July 2017, from https://www.ensembl.org
- Fagerberg, L., Hallström, B. M., Oksvold, P., Kampf, C., Djureinovic, D., Odeberg, J., … Uhlén, M. (2014). Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Molecular & Cellular Proteomics, 13, 397–406.10.1074/mcp.M113.035600
- Hayashi, K., Cao, T., Passmore, H., Jourdan-Le Saux, C., Fogelgren, B., Khan, S., … Csiszar, K. (2004). Progressive hair loss and myocardial degeneration in rough coat mice: Reduced lysyl oxidase-like (LOXL) in the skin and heart. Journal of Investigative Dermatology, 123, 864–871.10.1111/j.0022-202X.2004.23436.x
- Leiva, A. G., Chen, A. L., Devarajan, P., Chen, Z., Damanpour, S., Hall, J. A., … Wikramanayake, T. C. (2014). Loss of Mpzl3 function causes various skin abnormalities and greatly reduced adipose depots. Journal of Investigative Dermatology, 134, 1817–1827.10.1038/jid.2014.94
- Maksakova, I. A., Romanish, M. T., Gagnier, L., Dunn, C. A., van de Lagemaat, L. N., & Mager, D. L. (2006). Retroviral elements and their hosts: Insertional mutagenesis in the mouse germ line. PLoS Genetics, 2, e2.10.1371/journal.pgen.0020002
- McMahon, K. A., Hiew, S. Y., Hadjur, S., Veiga-Fernandes, H., Menzel, U., Price, A. J., … Brady, H. J. (2007). Mll has a critical role in fetal and adult hematopoietic stem cell self-renewal. Cell Stem Cell, 1, 338–345.10.1016/j.stem.2007.07.002
- Mouse ENCODE Consortium. (2014). A comparative encyclopedia of DNA elements in the mouse genome. Nature, 20, 355–364.
- Mouse Genome Database (MGD). (2017). Mouse genome database group: The mouse genome informatics website. Bar Harbor ME: The Jackson Laboratory. Retrieved July 2017, from https://www.informatics.jax.org
- Online Mendelian Inheritance in Man (OMIM). (2017). McKusick-Nathans institute of genetic medicine. Baltimore, MD: Johns Hopkins University. Retrieved July 2017, from https://omim.org
- Park, Y.-G., Hayasaka, S., Takagishi, Y., Inouye, M., Okumoto, M., & Oda, S. (2001). Histological characteristics of the pelage skin of rough fur mice (C3H/HeJ-ruf/ruf). Experimental Animals, 50, 179–182.10.1538/expanim.50.179
- Ramirez, F., Feliciano, A. M., Adkins, E. B., Child, K. M., Radden II, L. A., Salas, A., … King, T. R. (2013). The juvenile alopecia mutation (jal) maps to mouse Chromosome 2, and is an allele of GATA binding protein 3 (Gata3). BMC Genetics, 14, 40.10.1186/1471-2156-14-40
- Ruvinsky, I., Chertkov, O., Borue, X. V., Agulnik, S. I., Gibson-Brown, J. J., Lyle, S. R., & Silver, L. M. (2002). Genetics analysis of mouse mutations Abnormal feet and tail and rough coat, which cause developmental abnormalities and alopecia. Mammalian Genome, 13, 675–679.10.1007/s00335-002-2191-6
- Sen, G. L., Boxer, L. D., Webster, D. E., Bussat, R. T., Qu, K., Zarnegar, B. J., … Khavari, P. A. (2012). ZNF750 is a p63 target gene that induces KLF4 to drive terminal epidermal differentiation. Developmental Cell, 22, 669–677.10.1016/j.devcel.2011.12.001
- Sweet, H. O., Oda, S. I., Taylor, B. A., Rowe, L., Davisson, M. T., Cook, S., … Sundberg, J. (1990). Rough fur (ruf). Mouse Genome, 86, 236–237.
- Tang, T., Li, L., Tang, J., Li, Y., Lin, W. Y., Martin, F., … de Sauvage, F. J. (2010). A mouse knockout library for secreted and transmembrane proteins. Nature Biotechnology, 28, 749–755.10.1038/nbt.1644
- Thien, C. B., Scaife, R. M., Papadimitriou, J. M., Murphy, M. A., Bowtell, D. D., & Langdon, W. Y. (2003). A mouse with a loss-of-function mutation in the c-Cbl TKB domain shows perturbed thymocyte signaling without enhancing the activity of the ZAP-70 tyrosine kinase. The Journal of Experimental Medicine, 197, 503–513.10.1084/jem.20021498
- Wikramanayake, T. C., Borda, L. J., Kirsner, R. S., Wang, Y., Duffort, S., Reyes-Capo, A., … Perez, V. L. (2016). Loss of MPZL3 function causes seborrheic dermatitis-like phenotype in mice. Experimental Dermatology, 26, 736–738.
- Xu, X., Kedlaya, R., Higuchi, H., Ikeda, S., Justice, M. J., Setaluri, V., & Ikeda, A. (2010). Mutation in archain 1, a Subunit of COPI coatomer complex, causes diluted coat color and Purkinje cell degeneration. PLoS Genetics, 6, e1000956.10.1371/journal.pgen.1000956
