Abstract
The advent of genetically engineered systems, including transgenic animals and recombinant viral vectors, has facilitated a more detailed understanding of the molecular and cellular substrates regulating brain function. In this review we highlight some of the most recent molecular biology and genetic technologies in the experimental “systems neurosciences,” many of which are rapidly becoming a methodological standard, and focus in particular on those tools and techniques that permit the reversible and cell-type specific manipulation of neurons in behaving animals. These newer techniques encompass a wide range of approaches including conditional deletion of genes based on Cre/loxP technology, gene silencing using RNA interference, cell-type specific mapping or ablation and reversible manipulation (silencing and activation) of neurons in vivo. Combining these approaches with viral vector delivery systems, in particular adeno-associated viruses (AAV), has extended, in some instances greatly, the utility of these tools. For example, the spatially- and/or temporally-restricted transduction of specific neuronal cell populations is now routinely achieved using the combination of Cre-driver mice and stereotaxic-based delivery of AAV expressing Cre-dependent cassettes. We predict that the experimental application of these tools, including creative combinatorial approaches and the development of even newer reagents, will prove necessary for a complete understanding of the neuronal circuits subserving most neurobiological functions, including the regulation of sleep and wake.
Abbreviations
| AAV | = | adeno-associated viruses |
| GluClαβ | = | C. elegans glutamate- and ivermectin (IVM)-gated chloride channel subunits α and β |
| DOX | = | doxycycline |
| DREADD | = | designer receptors exclusively activated by designer drugs |
| EEG | = | electroencephalogram |
| MCH | = | melanin-concentrating hormone |
| NREM | = | non-rapid eye movement sleep |
| PG | = | Prostaglandin |
| REM | = | rapid eye movement sleep |
| RNAi | = | RNA interference |
| tTA | = | tetracycline-responsive transcription factor. |
Introduction
Three decades ago Francis Crick envisioned technologies that would permit the inactivation of specified neuronal populations, in turn enabling scientists to determine how “function follows structure” (of the brain). To this end, one of the more enduring mysteries in the neurosciences is the brain mechanisms and substrates (i.e., key circuit nodes, their transmitters and their targets) that regulate sleep – a highly conserved and vital biological process. Over the last 2 decades, researchers have developed a wide range of molecular and electrophysiological techniques and tools for probing and perturbing neural circuitry, including the circuitry that regulates sleep and wake in mammals. And while these tools and techniques have provided significant insight into the molecular and neurotransmitter systems used by the brain to regulate sleep and wake (cf. section ‘Neuronal mechanisms of sleep-wake regulation’), many of these approaches have non-trivial limitations, in particular with respect to data interpretation. For example, pharmacologic approaches such as receptor antagonists and protein inhibitors are often limited by low solubility, poor blood-brain-barrier permeability or other “off-target” side effects. Global knockout approaches have limited temporal and spatial resolution and can be confounded by ontogenetic issues. Even acute lesion approaches (including so-called “cell-specific” lesions) can produce collateral damage to adjacent brain structures that may, in turn, produce effects that are epiphenomenal to the lesion itself. Fortunately the emergence of newer conditional genomic models is helping to overcome many of these technical and interpretational concerns. Specific examples, which are discussed herein, include conditional deletion of genes based on Cre/loxP technology, gene silencing using RNA interference, cell-type specific mapping or ablation and genetically engineered receptor-channel systems (e.g. opsin-based optical switches, designer receptors exclusively activated by designer drugs and mutated non-mammalian channel systems), the latter of which permit the “remote” and reversible manipulation of neuronal activity in behaving animals. In addition to providing an overview of these emerging molecular biological techniques, we will provide literature-based examples of their application in experiments seeking a detailed understanding of the anatomic and molecular mechanism governing behavioral state regulation.
Models of sleep-wake regulation
Technical advances have often precipitated quantum leaps in our understanding of neurobiological processes. For example, Hans Berger's discovery in 1929 that electrical potentials recorded from the human scalp took the form of sinusoidal waves, the frequency of which was directly related to the level of wakefulness of the subject, led to rapid advances in our understanding of sleep-wake regulation, in both animals and humans alike. To this day the electroencephlogram (EEG), in conjunction with the electromyogram (i.e., electrical activity produced by skeletal muscles), represents the data “backbone” of nearly every experimental and clinical assessment that seeks to correlate behavior and physiology with the activity of cortical neurons in behaving animals, including humans. In most basic sleep research laboratories these EEG recordings are performed using a cable-based system wherein acquired data is subjected off-line to pattern and spectrum analysis (e.g. fast Fourier transform) to determine the vigilance state of the subject under recording ().Citation1,2,3 Over the years, and on the basis of EEG interpretation, several models of sleep-wake regulation, both circuit- and humoral-based, have been proposed ().
Figure 1. Sleep bioassay system for rodents. (A) To monitor electroencephalogram (EEG) signals, stainless steel screws are implanted epidurally over the frontal cortical and parietal areas of one hemisphere. In addition, electromyogram (EMG) activity is monitored by stainless steel, teflon-coated wires placed bilaterally within the trapezius muscles. (B) wakefulness (i) is characterized by low to moderate voltage EEG and the occurrence of EMG activity, whereas NREM sleep (ii) is identified by the appearance of large, slow brain waves with a rhythm below 0.5–4 Hz (orange frequencies in the fast Fourier transform, FFT, of the EEG) and REM sleep (iii), exhibits a shift back to a rapid low-voltage EEG and the appearance of brain waves in the theta range, i.e., 6–10 Hz (blue frequencies in FFT of the EEG).
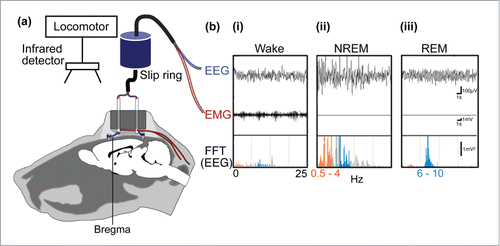
Figure 2. Circuit basis of sleep-wake regulation. Model 1 (shown in panel a): Adenosine inhibits the release of acetycholine from basal forebrain (BF) cholinergic neurons to produce slow-wave sleep. Model 2 (shown in panels b-d): a flip–flop switching mechanism involving mutually inhibitory interactions between sleep-promoting neurons in the ventrolateral preoptic area (VLPO) and wake-promoting neurons in the hypothalamus [i.e., histaminergic tuberomammilary nucleus (TMN)], and brainstem [i.e., noradrenergic locus coeruleus (LC), serotonergic dorsal raphe nucleus (DR), and cholinergic laterodorsal tegmental nucleus (LDT)]. The flip-flop switch between the VLPO and hypothalamus and brainstem is stabilized by orexin/hypocretin (OX/Hcrt) inputs. Adenosine is known to act as an endogenous somnogen and promotes sleep via inhibitory A1 receptors (A1) in the basal forebrain, VLPO and TMN and excitatory A2A receptors (A2A) in the nucleus accumbens (NAc) and VLPO.Citation98-101 Other Abbreviations: Ach, acetylcholine; 5-HT, serotonin, NE, norepinephrine.
![Figure 2. Circuit basis of sleep-wake regulation. Model 1 (shown in panel a): Adenosine inhibits the release of acetycholine from basal forebrain (BF) cholinergic neurons to produce slow-wave sleep. Model 2 (shown in panels b-d): a flip–flop switching mechanism involving mutually inhibitory interactions between sleep-promoting neurons in the ventrolateral preoptic area (VLPO) and wake-promoting neurons in the hypothalamus [i.e., histaminergic tuberomammilary nucleus (TMN)], and brainstem [i.e., noradrenergic locus coeruleus (LC), serotonergic dorsal raphe nucleus (DR), and cholinergic laterodorsal tegmental nucleus (LDT)]. The flip-flop switch between the VLPO and hypothalamus and brainstem is stabilized by orexin/hypocretin (OX/Hcrt) inputs. Adenosine is known to act as an endogenous somnogen and promotes sleep via inhibitory A1 receptors (A1) in the basal forebrain, VLPO and TMN and excitatory A2A receptors (A2A) in the nucleus accumbens (NAc) and VLPO.Citation98-101 Other Abbreviations: Ach, acetylcholine; 5-HT, serotonin, NE, norepinephrine.](/cms/asset/838c2a90-95f1-40fb-a23c-d7e55b412193/ktmp_a_1075095_f0002_c.gif)
Humoral mechanisms of sleep-wake regulation
The neural and cellular basis of sleep need or, alternatively, “sleep drive” remains unresolved, but has been conceptualized as a homeostatic pressure that builds during the waking period and is dissipated by sleep. One theory is that endogenous somnogenic factors accumulate during wake and that their gradual accumulation is the underpinning of sleep homeostatic pressure. And while the first formal hypothesis that sleep is regulated by humoral factors is credited to Rosenbaum in 1892,Citation4 it was IshimoriCitation5,6 and PieronCitation7 who independently, and over 100 years ago, demonstrated the existence of sleep-promoting chemicals. Both researchers proposed, and indeed proved, that hypnogenic substances or “hypnotoxins” were present in the cerebral spinal fluid (CSF) of sleep-deprived dogs.Citation8 Over the past century several additional putative hypnogenic substances implicated in the sleep homeostatic process have been identified (for review, seeCitation9), including prostaglandin (PG) D2Citation10 (for review, seeCitation11), cytokinesCitation12 (for review, seeCitation13), adenosineCitation14 (for review, seeCitation15), anandamide,Citation16 and the urotensin II peptide.Citation17 Additional clues into the “humoral mechanisms” underpinning sleep regulation have been gleaned from study of individuals exhibiting “sickness behavior”, i.e., the fever, malaise, increased pain sensitivity, anorexia, and changes in sleep–wake typically observed during a bacterial or viral infection (for review, seeCitation18). Sickness behavior links to a cascade of pro-inflammatory mediators, including a wide range of cytokines and PGs that trigger an array of physiological responses termed the acute phase reaction. It is now an accepted fact that sleep regulation and the production of pro-infammatory cytokines by the host defense (immune) system are strongly interrelated.Citation19-21 The sleep patterns of humans in response to elevated levels of cytokines are complicated and dose-dependent, and include increased or decreased non-rapid eye movement (NREM) sleep. Specifically, human studies have shown that increased levels of proinflammatory cytokines (at plasma concentrations capable of inducing a fever) disrupt, rather than promote, sleep. On the other hand, smaller increases in plasma concentrations of proinflammatory cytokines, such as occurs during the circadian cycle, can increase NREM sleep and cortical slow wave intensity.Citation22 By contrast, rodents treated with proinflammatory cytokines exhibit an enhancement of NREM sleep and a decrease in rapid eye movement (REM) sleep. Although cytokine production is inevitably accompanied by the secretion of PGs, the increase of NREM sleep during an infection is independent of PGs,Citation23 a surprising fact considering: 1) the fever response is completely dependent on PGE2 type EP3 receptor signalingCitation24,25 and 2) PGs have been implicated in the regulation of sleep.Citation11 How and why immune signaling molecules and other hypnogenic substances modulate sleep remain incompletely understood and are currently areas of active investigation.
Circuit mechanisms of sleep-wake regulation
Experimental work by Economo,Citation26-28 Ranson,Citation29 Moruzzi and Magoun,Citation30 and others in the early and mid 20th century produced findings that inspired circuit-based theories of sleep and wake and, to a certain degree, overshadowed the then prevailing humoral theory of sleep. To date, several “circuit models” have been proposed, each informed by data of varying quality and quantity (for review, seeCitation31-33). One model, for example, proposes that slow wave sleep is generated by adenosine-driven inhibition of acetylcholine release from cholinergic neurons in the basal forebrain (BF),Citation34 although several studies has shown that cholinergic BF neurons are not essential for sleep induction.Citation35,36 Another contemporary circuit model posits a flip-flop switching mechanism involving mutually inhibitory interactions between sleep-promoting neurons in the ventrolateral preoptic area (VLPO) and wake-promoting neurons in the brain stem and hypothalamus.Citation31,37,38 The flip-flop model further predicts that orexin-/hypocretin-containing neurons of the lateral hypothalamus provide a stabilizing influence on the switch, to prevent unwanted state transitions such as occurs in narcolepsy.Citation39-41 A similar mutually inhibitory interaction between the ventral periaqueductal gray, lateral pontine tegmentum, and sublaterodorsal nucleus in the brainstem has been proposed for the switching in and out of REM sleep.Citation42,43 And while these models have proven valuable heuristics and provided important interpretative frameworks for studies in sleep research, a fuller understanding of the circuits regulating sleep-wake will require a more complete knowledge of its components.
It is also the case that a unified model accounting for the myriad of state-dependent changes in physiology remains lacking. As an example, current models have (near uniformly) failed to address the regulated reduction in body temperature that occurs with sleep. Indeed, a reduction in core body temperature facilitates the entry into sleep and even modest changes to shell or core body temperature during sleep can positively or adversely affect sleep quality and consolidation,Citation44,45 suggesting a direct influence of thermosensory afferents on sleep circuits. Given moreover that the regulation of sleep and body temperature is temporally coordinated, it is likely, at least to a certain extent, that these systems share common neuronal circuits. In general support of this hypothesis, the preoptic forebrain contains neurons that are important for both sleep and temperature control.Citation46,47 And, similarly, the lateral parabrachial nucleus, which is linked with arousal control, receives peripheral cutaneous thermosensory signals.Citation48-50 Deconstructing these shared and distinct circuits will undoubtedly be facilitated by the development of newer molecular-genetic tools, and ulitimately should inform a unified model of sleep-wake regulation.
Molecular biology and genetic technologies in sleep research
As indicated, the combinatorial application of transgenic mice and viral vector delivery systems in systems-level neuroscience research has become increasingly common. Indeed, the power of this experimental approach is undeniable. And while a variety of viral vector systems have been harnessed for experimental purposes, we would highlight work using adeno-associated viruses (AAV) as these have enabled scientist to address long-standing questions in neurobiology. AAV are single-stranded DNA-containing parvoviruses of 57 serotypes classified in 7 speciesCitation51 that are currently not known to cause disease. Recombinant AAV vectors are replication deficient and AAV-delivered transgenes (typically no larger than 5 kb) mostly persist as highly stable, actively transcribed episomes enabling long-lasting gene expression in non-dividing cells. Although isolation of an AAV serotype 2 clone facilitated development of stable vectors for transgenic and therapeutic gene delivery, the level of gene expression in cells infected with AAV2 is generally low. As tissue specificity and affinity is determined by proteins that are present in the virus capsid (protein shell or viral envelope), successful attempts to pseudotype and engineer capsids have resulted in a great variety of AAV serotypes suitable for the infection of neurons in a wide range of animal species. For example, researchers can take advantage of AAV serotype variability to infect interdigitating populations of neurons in miceCitation52 or deliver transgenes to brains of vertebrates (e.g. zebra finches) that cannot be genetically modified by reproductive technologies.Citation53 Due to this great versatility, it is no surprise that AAV has become one of the most widely used biological tools of today's neuroscience.
Conditional gene manipulations based on Cre/loxP technology and focal RNA interference
Transgenic animals with constitutive gene disruptions have provided important insights into the in vivo roles of various genes (and their gene products) in sleep wake regulation. Interpretation of experimental data generated using constitutive gene disruption does, however, warrant caution given several limiting features of these animals, including: (i) approximately 15% of knockouts are developmentally lethal and so studies employing these mice are restricted to embryonic development, making it virtually impossible to relate the function of the deleted gene to sleep-wake; (ii) ontogenetic complications that result in abnormal development of other systems or compensatory alterations (e.g. levels of neurotransmitters or their receptors), which may contribute to the development of a phenotype that is epiphenomenal to the knockout itself; and (iii) constitutive knockout animals are limited in the ability to inform the localization of individual brain areas or neurons involved in sleep-wake regulation.
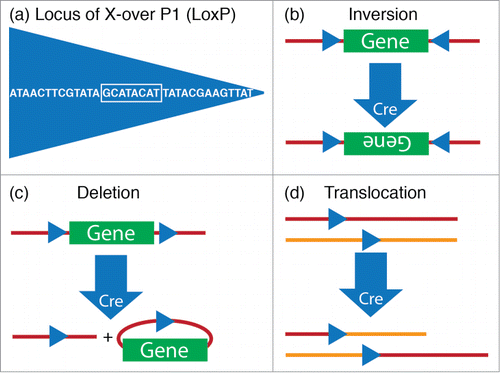
On the other hand, Cre or Flp recombinase mediated DNA recombination in genetically engineered mice, i.e., conditional knockouts, has proven a powerful approach for evaluating the role of genes, including “sleep genes”, in a spatially and temporally restricted manner.Citation54 Cre recombination was originally discovered in the P1 bacteriophage as part of the virus' life cycle.Citation55,56 The Cre enzyme recombines a pair of short target sequences called the lox sites, a mechanism which the P1 phage uses to circularize and facilitate replication of its genomic DNA during reproduction. Flp recombination is analogous to the Cre/lox system but involves recombination of DNA sequences flanked by FRT (short for “flippase recognition target”) sites derived from baker's yeast (Saccharomyces cerevisiae). For one reason or another, the Cre/lox recombination strategy has been the preferred recombination approach for the vast majority of transgenic manipulations in mice and other organisms.Citation57,58 And it is the orientation and location of the loxP sequences that determine whether Cre catalyzes a deletion, inversion, or chromosomal translocation of DNA sequences (). By genetic targeting of Cre (or Flp) to discrete populations of neurons and crossing the mice harboring these transgenes with mice bearing loxP (or FRT) modified alleles, it is possible to modulate these genes in a neuron-specific fashion.Citation59 Alternatively, AAV expressing either Cre or Flp can be stereotaxically-injected into specific nuclei in the brains of mice bearing loxP- or FRT-modified alleles to restrict gene expression to the site of AAV injection.Citation25,60 The utility of this technical approach was elegantly illustrated in 2 recent studies.Citation61,62 In the first study, Lazarus and colleagues employed, in combination, conditional A2A receptor knockout mice and stereotaxic-based microinjections of Cre-expressing AAV into the nucleus accumbens to show that A2A receptors in the nucleus accumbens promote sleep.Citation61 In the second study, Anaclet and colleagues placed microinjection of AAV-Cre into the parafacial zone of conditional vesicular GABA transporter mice to show that GABAergic neurons of the medullary parafacial zone are required for normal amounts of slow-wave-sleep.Citation62
Figure 3. Cre-lox system. (A) The 34-base pair loxP sequence consists of an 8-base pair core sequence where recombination takes place (white box) and 2 flanking 13-base pair inverted repeats. (B–D) The outcome of recombination by Cre, a P1 Bacteriophage enzyme that is encoded by the locus originally named as “causes recombination”, is determined by the location and orientation of lox sites. (B) Cre mediates the inversion of the floxed DNA segment, if lox sites are oriented in opposite directions. (C) Cre mediates a deletion of the floxed DNA segment, if lox sites are oriented in the same direction. (D) Cre mediates a translocation, if lox sites are located on different chromosomes.
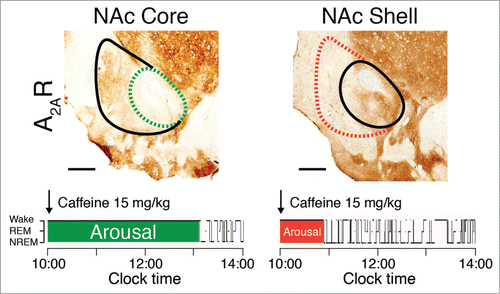
Also in the landmark Lazarus et al. paper on the arousal-promoting effect of caffeine,Citation61 the world's most widely prescribed psychoactive drug, the authors pioneered the use of RNA interference (RNAi) to silence focally the expression of A2A receptors in the brain of rats and show that deletion of A2A receptors in the shell of the nucleus accumbens is sufficient to abolish the arousal effect of caffeine (). RNAi is a system within living cells that helps regulate which genes are active and also the magnitude of their activity.Citation63 This gene regulation process also includes the interaction of small interfering RNA with mRNA to prevent mRNA from producing a protein. The RNAi can be applied in any animal model and by using local infection with AAV carrying short-hairpin RNA, focal RNAi can be produced in live animals in an efficient and cost-effective manner. Moreover, and in contrast to Cre/loxP conditional knockout models, rats or even more phylogenetically advanced organisms, such as non-human primates, which can provide a better approximation of human brain responses, can be used as experimental model systems.
Figure 4. The arousal effects of caffeine are abolished in rats with site-specific deletion of A2A receptors (A2AR) in the shell of the nucleus accumbens (NAc). To identify the neurons on which caffeine acts to produce arousal, A2A receptors were focally depleted by bilateral injections of adeno-associated virus carrying short-hairpin RNA for the A2A receptor into the core (dashed green line in the left panel) or shell (dashed red line in the right panel) of the NAc of rats.Citation61 Typical hypnograms that show changes in wakefulness and in rapid eye movement (REM) and non-REM (NREM) sleep after administration of caffeine at a dose of 15 mg/kg indicate that rats with a shell, but not a core, knockdown of the A2A receptors showed a strongly attenuated caffeine arousal. Green and red areas in the hypnograms represent wakefulness after caffeine administration that correspond to the depletion of A2A receptors in the respective core and shell of the NAc.
Many labs have also developed lines of mice with loxP-flanked sequences that disrupt expression of genes (“transcriptional disruptors”), effectively extending the utility of conditional knockouts by addressing some of their limitations. In their basal state, transcriptional disruptor mice typically demonstrate a phenotype identical to that of constitutive knockout mice; after exposure to Cre recombinase the loxP-flanked blocking sequence is removed and gene expression (and, typically, the phenotype) is normalized. Transcriptional disrupter mice are particularly useful in situations where the gene(s) of interest are more widely and diffusely expressed in the brain. In other words, gene re-expression can be targeted to discrete sites, which in turn greatly facilitates functional analysis. Focal gene reactivation using transcriptional disruptor mice also offers other potential advantages over traditional conditional knockout mice including: the ability to 1) perform a relatively quick anatomic survey for gene function in various brain regions across the neuraxis, and 2) normalize gene expression only in sites with a latent genetic capacity to express the gene of interest, i.e., eutopic expression. As an excellent experimental example of the gene “reactivation” approach, Scammell and colleagues placed focal injections of AAV containing Cre into discrete brain regions of mice with re-activatable orexin-2 receptors. In doing so, they found that normal expression and function of orexin-2 receptors, and hence orexin signaling, in the posterior hypothalamus, including the tuberomammilary nucleus, plays an essential role in the wake-promoting effects of orexins.Citation64
Conditional tracing of long axonal pathways and lesioning of neuronal cell populations
In addition to loxP-modified mice, investigators have also taken advantage of the large number of transgenic Cre-expressing mice available for use in a wide range of experiments, including the anterograde tracing of neural projections from cell-type specific neuronal populations. For example, Gautron and colleagues developed an AAV vector that encodes a humanized Renilla green fluorescent protein (hrGFP) whose expression is transcriptionally silenced by a neo cassette flanked by loxP sites.Citation65 This vector construct, which results in hrGFP protein expression only in neurons with Cre recombinase activity, was used in combination with leptin receptor-Cre mice to define the efferent projections of leptin-responsive neurons in the hypothalamus. In another example of this approach, stereotaxic microinjections of conditional hrGFP-AAV into the nucleus accumbens of mice in which Cre expression is driven by the promoter of the A2A receptor geneCitation66 were used to trace axonal projections of A2A receptor-positive neurons known to participate in sleep-wake regulation.
With little modifications, the same approach can readily be adapted for the lesioning of cell-type specific neuronal population. Toward this end we (Fuller and Lazarus) developed an AAV-based system for the transgenic expression of the highly cell-toxic fragment A of the diphtheria toxin (DTA-AAV). The original diphtheria toxin, an exotoxin secreted by Corynebacterium diphtheriae, is a single polypeptide consisting of 2 fragments A and B. Binding of fragment B to the cell surface allows fragment A to penetrate the host cell and act as a potent RNA translational inhibitor. In combination with mice expressing Cre under the control of the vesicular glutamate transporter 2, this transcriptionally silenced DTA-AAV was recently used by Saper and colleagues to demonstrate that glutamatergic neurons within the external lateral and lateral crescent subdivisions of the lateral pontine parabrachial nucleus critically contribute to hypercapnia-induced arousal.Citation49
As an alternative to using loxP-flanked neo cassettes as transcriptional stop sequences, transgenes are now commonly cloned into AAV plasmids in a double floxed inverted (FLEX) or double inverted orientation (DIO). Within the FLEX or DIO orientation, transgenes are cloned in reverse orientation, so that a nonsense transcript is produced until the transgene is exposed to Cre recombinase, which flips the gene into the sense orientation. The advantage to FLEX/DIO cassette is that it confers great selectivity, without transcriptional “leakage” in cells that lack Cre expression. We have, for example, recently generated a FLEX version of our DTA-AAV as well as adopted this system for all of our AAV-based genetically engineered receptor-channel systems (compare ‘Reversible in vivo silencing and activation of neurons in freely behaving animals’).
Reversible in vivo silencing and activation of neurons in freely behaving animals
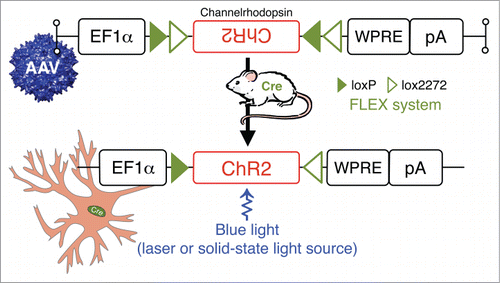
Significant research efforts have recently been directed at developing genetic-molecular tools to achieve reversible and cell-type specific in vivo silencing of neurons in awake, behaving animals. The obvious goal in developing these tools is to help establish a causal relationship between the activity of specific neurons (or neuronal populations) and behavioral and physiological outcomes. While several genetic tools have been developed for this purpose, including conditional blockade of neurotransmitter release and suppression of neuronal excitability, each method has distinct advantages as well as limitations. One tool that has been developed for acute and reversible in vivo silencing or activation of neurons is optogenetics technologies.Citation67 It would not be an exaggeration to state that optogenetics has ushered in a new era of neurobiology.Citation68 Simply put, an optically-driven “switch” can be placed into specific neurons of a living animal by the local microinjection of viral vectors expressing an opsin [e.g. excitatory channelrhodopsin-2 (ChR2), as shown in ] in a Cre-dependent configuration. Alternatively, the opsin gene can be placed under a cell-type specific promoter, limiting expression to, for example, only orexin-producing cells.Citation69 Transgenic mice have also been engineered to express opsins under various gene promoters. For example, Tsunematsu and colleagues generated transgenic mice in which halorhodopsin, which is orange light-driven chloride pump whose photoactivation results in the inhibition/silencing of neurons, was exclusively expressed in orexin neurons. In these halorhodopsin-based experiments, the authors showed that silencing of orexin neurons induced NREM sleep, which confirmed a role for orexin neurons in the maintenance of wakefulness.Citation70 It is also the case that optogenetic tools can be used to identify functional synaptic connectivity between specific neuronal populations, both in vivo and in brain slices. Originally coined “ChR2-assisted circuit mapping,” or CRACM for short, this technique involves combining direct photostimulation of presynaptic ChR2-expressing axons/terminals with patch-clamp recordings of post-synpatic neurons. In this arrangement, specific inputs are activated to evoke neurotransmitter release and establish functional synaptic connectivity. This elegant technique was recently used by Arrigoni and colleagues to show that release of histamine from neurons of the tuberomammilllary nucleus (TMN) can disinhibit the TMN and suppress (indirectly) the activity of sleep-active VLPO neurons to promote TMN neuronal firing, a finding that lends credence to the sleep-wake “flip-flop switch” hypothesis.Citation71 Creative variants of this technique, including the combined application of CRACM and retrogradely transported microspheres, have enabled the mapping of circuits spanning 3 synaptically-coupled sites within the brain.Citation72
Figure 5. Photostimulation of neurons in behaving animals: combining Cre-driver mice and stereotaxic-based delivery of adeno-associated virus (AAV) expressing Cre-dependent channelrhodopsin (ChR2). ChR2 are nonspecific cation channels, conducting H+, Na+, K+, and Ca2+ ions. ChR2 absorbs blue light with an absorption spectrum maximum at 480 nm resulting in the opening of a pore in the trans-membrane protein and depolarization of neurons by allowing for the flow of ions according to their electrochemical gradient.Citation102,103 Other Abbreviations: Cre, Cre recombinase; EF1α, archaeal elongation factor 1 α; loxP, locus of X-over P1; lox2272, variant of loxP; pA, poly A tail; WRPE, woodchuck hepatitis virus posttranscriptional regulatory element.
We next highlight 3 recently published (and related) studies in which optogenetic-based approaches were used to interrogate the neuronal circuitry subserving the regulation of REM and NREM sleep, with a particular emphasis on the role of lateral hypothalamic melanin-concentrating hormone (MCH) neurons.Citation73 In the first, Adamatidis and colleagues used AAV to deliver Cre-dependent light-actviated opsins (ChETA and halorhopdsin) within the lateral hypothalamus of mice expressing Cre recombinase under the prepro-MCH promoter. In vivo activation and inhibition of these neurons revealed that these neurons are critical for maintenance of REM sleep,Citation74 and that this links to GABA release onto TMN neurons by MCH neurons. The second study, by Shiromani and colleagues, also employed an AAV-based delivery approach, but instead used a MCH promoter system to drive expression of channelrhodposin in MCH neurons of wildtype mice.Citation75 Optogenetic stimulation of the ChR2-expessing MCH neurons during normal waking time reduced the length of waking bouts and increased both NREM and REM sleep. In the third, Tsunematsu and colleagues used a genetic approach that faciliated the temporal and spatial control of the expression of opsins and other transgenes.Citation76 They employed a tetracycline-controlled transcriptional activation technique (), which is a method of inducible expression wherein transcription depends on the tetracycline-responsive transcription factor (tTA) and is turned off at the TetO promoter in the presence of the antibiotic tetracycline [or its derivative, doxycycline (DOX); “Tet-Off”].Citation77,78 Initially, Tsunematsu and colleagues expressed ChR2 (E128T/T159C) in MCH neurons by generating mice with 2 transgenes in which the first transgene provides pro-MCH promoter-driven expression of tTA and the second enables tTA-dependent expression of ChR2.Citation76 Activation of MCH neurons in these mice increased time in REM sleep, whereas optogenetic inhibition, which was achieved through expression of tTA-dependent archaerhodopsinT,Citation79 did not affect the amount of REM sleep. Additionally, mice with genetically ablated MCH neurons (Tet-Off-controlled expression of DTA) showed an increase in wake and a decrease in NREM sleep without affecting REM sleep amount. Taken the together, the results from these 3 optogenetic-based studies suggest that MCH neurons contributes to both NREM and REM sleep, possibly in a state- and time-of-day dependent manner.
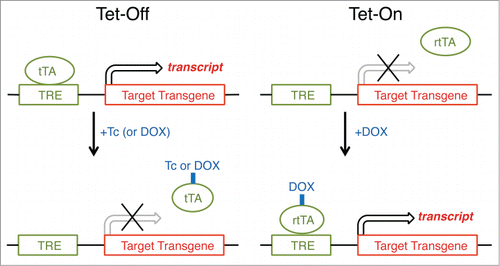
Figure 6. Inducible gene expression by using tetracycline-controlled transcriptional activation (Tet expression systems). Gene transcription is reversibly turned on or off in the presence of the antibiotic tetracycline (Tc) or doxycycline (DOX), a more stable tetracycline analog. In a Tet-Off system, tetracycline and its derivatives bind transactivator protein (tTA) and render it incapable of binding to the tetracycline response element (TRE) consisting of several TetO sequences and a minimal promoter, thereby preventing transcription of TRE-controlled genes. A Tet-On system works similarly, but the rtTA protein is capable of binding to the TRE operator, and hence initiating transcription of the transgene, only when bound by DOX. For most purposes, there is no inherent advantage of using the Tet-Off system over the Tet-On system, although there is no apparent literature example in which the Tet-Off system has been used to study the regulation and function of sleep.
It is worth noting that, despite its undeniable contribution to the experimental neurosciences, in vivo optogenetic tools are not without limiting features. These limitations include invasive instrumentation, the challenge of light penetration to larger brain regions, scalability issues, lower throughput, artificial synchronized patterns of activation/inhibition and limited direct evidence that photo-evoked release of neuropeptides is possible. And because of these limiting features, our labs and others have begun exploring and developing alternative tools for achieving in vivo, reversible silencing that also offers ease of implementation, no cabling into the CNS and a ligand that can be delivered peripherally or even in the drinking water. One such system was first introduced in 2007 by Andersen and colleagues,Citation80 and involves the AAV-based delivery of 2 channel subunits (α and β) that comprise a modified C. elegans glutamate- and ivermectin (IVM)-gated chloride channel (GluClαβ). This heteromeric channel prevents action potentials from firing by hyperpolarizing the membrane in a ligand-dependent manner. In this elegant proof-of-concept study, the authors showed that that the GluClαβ channel can be stably expressed in vivo without neurotoxicity, that this channel can be activated by (dose-dependent) systemic administration of IVM in vivo (at dosages that do not cause organismal toxicity), channel activation is reversible in vivo and, finally, that the channel-IVM mediated neuronal silencing can be used to manipulate behavior in awake, behaving animals. In a more recent study in sleep biology, the technique was instrumental in defining the neural substrates of emotion-driven cataplexy.Citation81 Here, Oishi and colleagues showed that the GluClαβ-IVM-mediated inhibition of the medial prefrontal cortex (mPFC) prevented chocolate-induced cataplexy in orexin knock-out mice, a finding that suggests a key role for the mPFC in positive emotions that trigger cataplexy.
Another recently developed system by Roth and colleagues permits the selective and “remote” manipulation (activation and silencing) of neuronal activity via all 3 major GPCR signaling pathways (Gi, Gs and Gq). These so-called “designer receptors exclusively activated by designer drugs” (DREADD) involve, broadly speaking, mutant GPCRs that do not respond to their endogenous ligands but are responsive to otherwise inert biological compounds (). The usefulness of these chemogenetic systems has been shown in various studies across the neurosciences and other fields.Citation82-85 In an important proof-of-concept study for sleep biology, the laboratory of Takeshi Sakurai demonstrated that DREADD-driven changes in the activity of orexin neurons can alter behavioral state.Citation86 More specifically, Gq-DREADD excitation of orexin neurons increased the amount of time spent in wakefulness, whereas Gi-DREADD inhibition of orexin neurons promoted NREM sleep. In related work, Inutsuka and colleagues activated orexin neurons, also using Gq-DREADD,Citation87 and observed increases in food and water consumption as well as locomotor activity and metabolic rate, suggesting that orexin neurons also contribute to the regulation of energy homeostasis. More recently, Anaclet and colleagues generated and experimentally deployed Cre-dependent versions of the Gq and Gi DREADD-AAV systems to establish necessity and sufficiency of a node of GABAergic brainstem neurons in generating slow-wave-sleep and cortical slow-wave-activity.Citation72
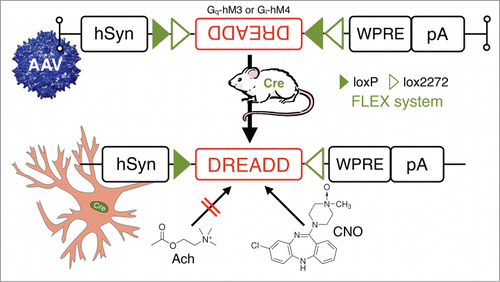
Figure 7. In vivo chemogenetic inhibition or activation of neurons in behaving animals: combining Cre-driver mice and stereotaxic-based delivery of adeno-associated virus (AAV) expressing Cre-dependent “designer receptor exclusively activated by designer drugs” (DREADD). DREADD permit temporal control of excitatory or inhibitory G-protein coupled receptor signaling in vivo by utilizing mutated human muscarinic acetylcholine (AcH) receptors.Citation82,83 These AcH receptors, excitatory hM3 and inhibitory hM4, are unresponsive to their natural ligand acetylcholine, but can be activated by nanomolar doses of the synthetic small-molecule clozapine-N-oxide (CNO). Other Abbreviations: Cre, Cre recombinase; hsyn, human synapsin promoter; loxP, locus of X-over P1; lox2272, variant of loxP; pA, poly A tail; WRPE, woodchuck hepatitis virus posttranscriptional regulatory element.
Newer generation DREADD systems are under development with several examples appearing in the recent literature. For example, a new Gi-coupled DREADD that uses the kappa-opioid receptor as a template (KORD) and is activated by the pharmacologically inert ligand salvinorin B was recently described.Citation88 Co-expression of the KORD and the Gq-coupled M3-DREADD within the same neuronal population permits the sequential and bidirectional control of the target neuronal population, and hence behavior.
Increasingly, the experimental framework in which both opto- and chemo-genetic techniques are being applied involves concurrent electrophysiologic and imaging techniques, such as tetrode recordingCitation89 or deep-brain fiber-optic endomicroscopy,Citation90 in behaving mice. Such innovative combinatorial approaches permit, for example, the simultaneous recording and perturbation of genetically defined sets of neurons, even in regions of high cellular heterogenecity. Hence the combined application of genetically driven system with in vivo electrophysiologic and/or imaging techniques will likely prove instrumental in elucidating the detailed circuit and synaptic basis of wake-sleep control.”
Reversible neurotransmission blocking by the tetanus neurotoxin and tetracycline-controlled transcriptional activation
Tetanus neurotoxin cleaves the synaptic vesicle-associated membrane protein and thus blocks vesicle-mediated neurotransmission.Citation91 Local reversible silencing of neurotransmission can be achieved by the injection of tTA-expressing AAV into transgenic mice with tetanus neurotoxin expression under the control of tTA. Such an approach has recently been used to genetically dissect the circuit-function relationships within the basal ganglia.Citation92 In this study, an AAV-tTA/tetanus neurotoxin approach was used to evaluate the functional roles of the direct (striatonigral) and indirect (striatopallidal) pathways in learning behaviors. Here the authors exploited the fact that substance P and enkephalin are selectively expressed in the direct or indirect pathway, respectively, and thus, AAV-mediated expression of tTA under the control of the substance P or enkephalin promoter induced specific tetanus neurotoxin-blocking of the striatonigral or striatopallidal neurons. In doing so, this study revealed that dopamine/D2R action on the indirect pathway is important for aversive, but not reward-based, learning. This same approach may also prove useful in determining the extent to which the 2 efferent pathways of the basal ganglia are required for the regulation of wakefulness.
The Tet-off system has also been widely used in combination with genetically engineered receptor-channel systems including opsins or DREADDs. One particularly useful application of the Tet-Off system is the generation of synthetic activity-dependent transgene traces in the brains of mice in which, for example, ChR2 or Gq-DREADD are expressed in a behavior-dependent manner through c-fos (a marker of neuronal activity) promoter-driven tTA expression.Citation93-95 To this end, a Tet-tagged version of the excitatory DREADD was recently used to selectively map and reactivate neuronal ensembles in the preoptic hypothalamus that were activated by the α2 adrenergic receptor agonist dexmedetomidine.Citation96 In doing so, the authors were able to demonstrate that dexmedetomidine–induced sedation is achieved, in part, through engagement of sleep-promoting hypothalamic circuitry.
Concluding remarks
It is the authors' surmise that the experimental application of the tools described in this review, including creative combinational applications, will prove critical to the process of elaborating the spatial and temporal properties of the circuitry mediating the transition between sleep and waking states, as well as developing a unified model for the humoral and neural mechanisms governing sleep-wake regulation. We also feel that “systems-level” sleep research will be greatly informed by large-scale gene network studies that employ functional genomics approaches such as transcriptome, proteome, and metabolome analysis for identifying “new targets” that might form the basis for the development of additional conditional transgenics. Indeed, the methods described herein or elsewhereCitation97 will not only make it possible to determine the detailed anatomic and molecular bases of sleep-wake regulation, but should also help to shed light on some of the greatest mysteries in systems somnology, including: “why do we sleep” and “what is the function of sleep?.” Disrupted sleep, including its voluntary loss and sleep disorders, are linked to traffic and work-related accidents as well as significant social losses due to an increased prevalence of mood and other neuropsychiatric disorders. Insufficient sleep is also an established independent risk factor for cardiovascular and metabolic diseases, such as diabetes and obesity, and is linked with increased cancer risk. Thus, while sleep has been a perpetual topic of scientific inquiry that continues to attract many scientists, it is also an important field that will greatly benefit society through the development of strategies to remedy sleep disorders and associated diseases.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
About the Authors
Patrick M Fuller is a principal investigator at Harvard Medical School and Beth Israel Deaconess Medical Center. The investigative focus of his laboratory is the cellular and synaptic basis by which the brain regulates sleep and wakeful consciousness. His experiments seek to link the activity of defined sets of neurons with neurobehavioral and electroencephalographic outcomes in behaving animals.

Akihiro Yamanaka is a principle investigator at the Research Institute of Environmental Medicine at Nagoya University. His experimental work has focused on orexin/hypocretin neurons since the peptide was first discovered in 1998. Using slice patch-clamp recording and optogenetics, his laboratory generated key insights into the regulation of sleep/wakefulness.

Michael Lazarus is a principal investigator at the International Institute for Integrative Sleep Medicine at the University of Tsukuba. He has made key contributions to our understanding of how prostaglandin and adenosine receptors in the brain regulate body temperature and sleep. His laboratory uses innovative genetically engineered systems to investigate homeodynamics in sleep and body temperature.

Funding
This work was supported by Japan Society for the Promotion of Science Grants-in-Aid for Scientific Research (B) 24300129 (to M.L.) and 23300142 (to A.Y.); World Premier International Research Center Initiative (WPI) from the Ministry of Education, Culture, Sports, Science, and Technology (to M.L.); the National Institutes of Health (NS26837 to P.M.F.); and a grant from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan (Grant-in-Aid for Scientific Research on Innovative Areas “Mesoscopic Neurocircuitry”, 23115103, to A.Y.).
References
- Kohtoh S, Taguchi Y, Matsumoto N, Wada M, Huang Z-L, Urade Y. Algorithm for sleep scoring in experimental animals based on fast Fourier transform power spectrum analysis of the electroencephalogram. Sleep Biol Rhythms 2008; 6:163-71; http://dx.doi.org/10.1111/j.1479-8425.2008.00355.x
- Tobler I, Deboer T, Fischer M. Sleep and Sleep Regulation in Normal and Prion Protein-Deficient Mice. J Neurosci 1997; 17:1869-79; PMID:9030645
- Oishi Y, Takata Y, Urade Y, Lazarus M. Polygraphic recordings to measure sleep in mice. J Vis Exp 2015: in press.
- Rosenbaum E. Warum müssen wir schlafen? : eine neue Theorie des Schlafes. Berlin: August Hirschwald, 1892.
- Kubota K. Kuniomi Ishimori and the first discovery of sleep-inducing substances in the brain. Neurosci Res 1989; 6:497-518; PMID:2677843; http://dx.doi.org/10.1016/0168-0102(89)90041-2
- Ishimori K. True cause of sleep: a hypnogenic substance as evidenced in the brain of sleep-deprived animals. Tokyo Igakkai Zasshi 1909; 23:429-57
- Legendre R, Pieron H. Recherches sur le besoin de sommeil consécutif à une veille prolongée. Z Allegem Physiol 1913; 14:235-62
- Inoué S, Honda K, Komoda Y. Sleep as neuronal detoxification and restitution. Behav Brain Res 1995; 69:91-6; http://dx.doi.org/10.1016/0166-4328(95)00014-K
- Urade Y, Hayaishi O. Prostaglandin D2 and sleep/wake regulation. Sleep Med Rev 2011; 15:411-8; PMID:22024172; http://dx.doi.org/10.1016/j.smrv.2011.08.003
- Ueno R, Ishikawa Y, Nakayama T, Hayaishi O. Prostaglandin D2 induces sleep when microinjected into the preoptic area of conscious rats. Biochem Biophys Res Commun 1982; 109:576-82; PMID:6960896; http://dx.doi.org/10.1016/0006-291X(82)91760-0
- Urade Y, Lazarus M. Prostaglandin D2 in the regulation of sleep. In: Shaw PJ, Tafti M, Thorpy MJ, eds. The Genetic Basis of Sleep and Sleep Disorders. New York: Cambridge University, 2013:73-83.
- Krueger JM, Walter J, Dinarello CA, Wolff SM, Chedid L. Sleep-promoting effects of endogenous pyrogen (interleukin-1). Am J Physiol 1984; 246:R994-9; PMID:6611091
- Krueger JM, Clinton JM, Winters BD, Zielinski MR, Taishi P, Jewett KA, Davis CJ. Involvement of cytokines in slow wave sleep. Prog Brain Res 2011; 193:39-47; PMID:21854954; http://dx.doi.org/10.1016/B978-0-444-53839-0.00003-X
- Porkka-Heiskanen T, Strecker RE, Thakkar M, Bjorkum AA, Greene RW, McCarley RW. Adenosine: a mediator of the sleep-inducing effects of prolonged wakefulness. Science 1997; 276:1265-8; PMID:9157887; http://dx.doi.org/10.1126/science.276.5316.1265
- Porkka-Heiskanen T, Kalinchuk AV. Adenosine, energy metabolism and sleep homeostasis. Sleep Med Rev 2011; 15:123-35; PMID:20970361; http://dx.doi.org/10.1016/j.smrv.2010.06.005
- Garcia-Garcia F, Acosta-Pena E, Venebra-Munoz A, Murillo-Rodriguez E. Sleep-inducing factors. CNS Neurol Disord Drug Targets 2009; 8:235-44; PMID:19689305; http://dx.doi.org/10.2174/187152709788921672
- Huitron-Resendiz S, Kristensen MP, Sánchez-Alavez M, Clark SD, Grupke SL, Tyler C, Suzuki C, Nothacker H-P, Civelli O, Criado JR, et al. Urotensin II Modulates Rapid Eye Movement Sleep through Activation of Brainstem Cholinergic Neurons. J Neurosci 2005; 25:5465-74; PMID:15944374; http://dx.doi.org/10.1523/JNEUROSCI.4501-04.2005
- Saper CB, Romanovsky AA, Scammell TE. Neural circuitry engaged by prostaglandins during the sickness syndrome. Nat Neurosci 2012; 15:1088-95; PMID:22837039; http://dx.doi.org/10.1038/nn.3159
- Krueger JM, Obal F, Jr, Fang J, Kubota T, Taishi P. The Role of Cytokines in Physiological Sleep Regulation. Ann NY Acad Sci 2001; 933:211-21; PMID:12000022; http://dx.doi.org/10.1111/j.1749-6632.2001.tb05826.x
- Krueger JM, Majde JA. Humoral Links between Sleep and the Immune System: Research Issues. Ann NY Acad Sci 2003; 992:9-20; PMID:12794042
- Mullington JM, Hinze-Selch D, Pollmacher T. Mediators of inflammation and their interaction with sleep: relevance for chronic fatigue syndrome and related conditions. Ann NY Acad Sci 2001; 933:201-10; PMID:12000021; http://dx.doi.org/10.1111/j.1749-6632.2001.tb05825.x
- Mullington J, Korth C, Hermann DM, Orth A, Galanos C, Holsboer F, Pollmächer T. Dose-dependent effects of endotoxin on human sleep. Am J Physiol Regul Integr Comp Physiol 2000; 278:R947-55; PMID:10749783
- Oishi Y, Yoshida K, Scammell TE, Urade Y, Lazarus M, Saper CB. The roles of prostaglandin E2 and D2 in lipopolysaccharide-mediated changes in sleep. Brain Behav Immun 2015; 47:172-7; PMID:25532785; http://dx.doi.org/10.1016/j.bbi.2014.11.019
- Ushikubi F, Segi E, Sugimoto Y, Murata T, Matsuoka T, Kobayashi T, Hizaki H, Tuboi K, Katsuyama M, Ichikawa A, et al. Impaired febrile response in mice lacking the prostaglandin E receptor subtype EP3. Nature 1998; 395:281-24; PMID:9751056; http://dx.doi.org/10.1038/26233
- Lazarus M, Yoshida K, Coppari R, Bass CE, Mochizuki T, Lowell BB, Saper CB. EP3 prostaglandin receptors in the median preoptic nucleus are critical for fever responses. Nat Neurosci 2007; 10:1131-3; PMID:17676060; http://dx.doi.org/10.1038/nn1949
- Wilkins RH, Brody IA. ENcephalitis lethargica. Arch Neurol 1968; 18:324-8; PMID:4868286; http://dx.doi.org/10.1001/archneur.1968.00470330114013
- von Economo C. Die Encephalitis lethargica. Wien Klin Wochenschr 1917; 30:581-5
- Economo CV. Sleep as a problem of localization. J Nerv Ment Dis 1930; 71:249-59; http://dx.doi.org/10.1097/00005053-193003000-00001
- Ranson SW. SOmnolence caused by hypothalamic lesions in the monkey. Arch Neurol Psychiatry 1939; 41:1-23; http://dx.doi.org/10.1001/archneurpsyc.1939.02270130011001
- Moruzzi G, Magoun HW. Brain stem reticular formation and activation of the EEG. Electroencephalogr Clin Neurophysiol 1949; 1:455-73; PMID:18421835; http://dx.doi.org/10.1016/0013-4694(49)90219-9
- Saper CB, Fuller PM, Pedersen NP, Lu J, Scammell TE. Sleep State Switching. Neuron 2010; 68:1023-42; PMID:21172606; http://dx.doi.org/10.1016/j.neuron.2010.11.032
- Brown RE, Basheer R, McKenna JT, Strecker RE, McCarley RW. Control of Sleep and Wakefulness. Physiol Rev 2012; 92:1087-187; PMID:22811426; http://dx.doi.org/10.1152/physrev.00032.2011
- Fuller PM, Gooley JJ, Saper CB. Neurobiology of the Sleep-Wake Cycle: Sleep Architecture, Circadian Regulation, and Regulatory Feedback. J Biol Rhythms 2006; 21:482-93; PMID:17107938; http://dx.doi.org/10.1177/0748730406294627
- Jones BE. Activity, modulation and role of basal forebrain cholinergic neurons innervating the cerebral cortex. Prog Brain Res 2004;145:157-69; PMID: 14650914; http://dx.doi.org/10.1016/S0079-6123(03)45011-5
- Blanco-Centurion C, Xu M, Murillo-Rodriguez E, Gerashchenko D, Shiromani AM, Salin-Pascual RJ, Hof PR, Shiromani PJ. Adenosine and Sleep Homeostasis in the Basal Forebrain. J Neurosci 2006; 26:8092-100; PMID:16885223; http://dx.doi.org/10.1523/JNEUROSCI.2181-06.2006
- Kapás L, Obál Jr F, Book AA, B. Schweitzer J, Wiley RG, Krueger JM. The effects of immunolesions of nerve growth factor-receptive neurons by 192 IgG-saporin on sleep. Brain Res 1996; 712:53-9; http://dx.doi.org/10.1016/0006-8993(95)01431-4
- Saper CB, Scammell TE, Lu J. Hypothalamic regulation of sleep and circadian rhythms. Nature 2005; 437:1257-63; PMID:16251950; http://dx.doi.org/10.1038/nature04284
- Fort P, Bassetti CL, Luppi PH. Alternating vigilance states: new insights regarding neuronal networks and mechanisms. Eur J Neurosci 2009; 29:1741-53; PMID:19473229; http://dx.doi.org/10.1111/j.1460-9568.2009.06722.x
- Lin L, Faraco J, Li R, Kadotani H, Rogers W, Lin X, Qiu X, de Jong PJ, Nishino S, Mignot E. The sleep disorder canine narcolepsy is caused by a mutation in the hypocretin (orexin) receptor 2 gene. Cell 1999; 98:365-76; PMID:10458611; http://dx.doi.org/10.1016/S0092-8674(00)81965-0
- Chemelli RM, Willie JT, Sinton CM, Elmquist JK, Scammell T, Lee C, Richardson JA, Williams SC, Xiong Y, Kisanuki Y, et al. Narcolepsy in orexin knockout mice: molecular genetics of sleep regulation. Cell 1999; 98:437-51; PMID:10481909; http://dx.doi.org/10.1016/S0092-8674(00)81973-X
- Thannickal TC, Moore RY, Nienhuis R, Ramanathan L, Gulyani S, Aldrich M, Cornford M, Siegel JM. Reduced number of hypocretin neurons in human narcolepsy. Neuron 2000; 27:469-74; PMID:11055430; http://dx.doi.org/10.1016/S0896-6273(00)00058-1
- Lu J, Sherman D, Devor M, Saper CB. A putative flip–flop switch for control of REM sleep. Nature 2006; 441:589-94; PMID:16688184; http://dx.doi.org/10.1038/nature04767
- Fuller PM, Saper CB, Lu J. The pontine REM switch: past and present. J Physiol (Lond) 2007; 584:735-41; PMID:17884926; http://dx.doi.org/10.1113/jphysiol.2007.140160
- Murphy PJ, Campbell SS. Nighttime drop in body temperature: a physiological trigger for sleep onset? Sleep 1997; 20:505-11; PMID:9322266
- Raymann RJ, Swaab DF, Van Someren EJ. Skin deep: enhanced sleep depth by cutaneous temperature manipulation. Brain 2008; 131:500-13; PMID:18192289; http://dx.doi.org/10.1093/brain/awm315
- Romanovsky AA. Thermoregulation: some concepts have changed. Functional architecture of the thermoregulatory system. Am J Physiol Regul Integr Comp Physiol 2007; 292:R37-46; PMID:17008453; http://dx.doi.org/10.1152/ajpregu.00668.2006
- Szymusiak R, McGinty D. Hypothalamic regulation of sleep and arousal. Ann NY Acad Sci 2008; 1129:275-86; PMID:18591488; http://dx.doi.org/10.1196/annals.1417.027
- Fuller P, Sherman D, Pedersen NP, Saper CB, Lu J. Reassessment of the structural basis of the ascending arousal system. J Comp Neurol 2011; 519:3817-; http://dx.doi.org/10.1002/cne.22781
- Kaur S, Pedersen NP, Yokota S, Hur EE, Fuller PM, Lazarus M, Chamberlin NL, Saper CB. Glutamatergic Signaling from the Parabrachial Nucleus Plays a Critical Role in Hypercapnic Arousal. J Neurosci 2013; 33:7627-40; PMID:23637157; http://dx.doi.org/10.1523/JNEUROSCI.0173-13.2013
- Nakamura K, Morrison SF. A thermosensory pathway that controls body temperature. Nat Neurosci 2008; 11:62-71; PMID:18084288; http://dx.doi.org/10.1038/nn2027
- Chen R-F, Lee C-Y. Adenoviruses Types, Cell Receptors and Local Innate Cytokines in Adenovirus Infection. Int Rev Immunol 2014; 33:45-53; PMID:24127823; http://dx.doi.org/10.3109/08830185.2013.823420
- Lopez-Huerta V, Nakano Y, Bausenwein J, Jaidar O, Lazarus M, Cherassse Y, Garcia-Munoz M, Arbuthnott G. The neostriatum: two entities, one structure? Brain Struct Funct 2015; PMID:25652680; http://dx.doi.org/10.1007/s00429-015-1000-4
- Yazaki-Sugiyama Y, Yanagihara S, Fuller PM, Lazarus M. Acute inhibition of a cortical motor area impairs vocal control in singing zebra finches. Eur J Neurosci 2014: 41(1):97-108; PMID:25354166; http://dx.doi.org/10.1111/ejn.12757
- Lewandoski M. Conditional control of gene expression in the mouse. Nat Rev Genet 2001; 2:743-55; PMID:11584291; http://dx.doi.org/10.1038/35093537
- Sauer B, Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc Natl Acad Sci USA 1988; 85:5166-70; PMID:2839833; http://dx.doi.org/10.1073/pnas.85.14.5166
- Sternberg N, Hamilton D, Hoess R. Bacteriophage P1 site-specific recombination: II. Recombination between loxP and the bacterial chromosome. J Mol Biol 1981; 150:487-507; PMID:6276558; http://dx.doi.org/10.1016/0022-2836(81)90376-4
- Nagy A. Cre recombinase: The universal reagent for genome tailoring. Genesis 2000; 26:99-109; PMID:10686599; http://dx.doi.org/10.1002/(SICI)1526-968X(200002)26:2%3c99::AID-GENE1%3e3.0.CO;2-B
- Araki K, Imaizumi T, Okuyama K, Oike Y, Yamamura K-I. Efficiency of Recombination by Cre Transient Expression in Embryonic Stem Cells: Comparison of Various Promoters. J Biochem 1997; 122:977-82; PMID:9443813; http://dx.doi.org/10.1093/oxfordjournals.jbchem.a021860
- Tronche F, Kellendonk C, Kretz O, Gass P, Anlag K, Orban PC, Bock R, Klein R, Schutz G. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet 1999; 23:99-103; PMID:10471508; http://dx.doi.org/10.1038/12703
- Coppari R, Ichinose M, Lee CE, Pullen AE, Kenny CD, McGovern RA, Tang V, Liu SM, Ludwig T. The hypothalamic arcuate nucleus: A key site for mediating leptin's effects on glucose homeostasis and locomotor activity. Cell Metabolism 2005; 1:63-72; PMID:16054045; http://dx.doi.org/10.1016/j.cmet.2004.12.004
- Lazarus M, Shen H-Y, Cherasse Y, Qu W-M, Huang Z-L, Bass CE, Winsky-Sommerer R, Semba K, Fredholm BB, Boison D, et al. Arousal Effect of Caffeine Depends on Adenosine A2A Receptors in the Shell of the Nucleus Accumbens. J Neurosci 2011; 31:10067-75; PMID:21734299; http://dx.doi.org/10.1523/JNEUROSCI.6730-10.2011
- Anaclet C, Lin J-S, Vetrivelan R, Krenzer M, Vong L, Fuller PM, Lu J. Identification and Characterization of a Sleep-Active Cell Group in the Rostral Medullary Brainstem. J Neurosci 2012; 32:17970-6; PMID:23238713; http://dx.doi.org/10.1523/JNEUROSCI.0620-12.2012
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998; 391:806-11; PMID:9486653; http://dx.doi.org/10.1038/35888
- Mochizuki T, Arrigoni E, Marcus JN, Clark EL, Yamamoto M, Honer M, Borroni E, Lowell BB, Elmquist JK, Scammell TE. Orexin receptor 2 expression in the posterior hypothalamus rescues sleepiness in narcoleptic mice. Proc Natl Acad Sci USA 2011; 108:4471-6; PMID:21368172; http://dx.doi.org/10.1073/pnas.1012456108
- Gautron L, Lazarus M, Scott MM, Saper CB, Elmquist JK. Identifying the efferent projections of leptin-responsive neurons in the dorsomedial hypothalamus using a novel conditional tracing approach. J Comp Neurol 2010; 518:2090-108; PMID:20394060; http://dx.doi.org/10.1002/cne.22323
- Zhang J, Xu Q, Yuan X, ChÈrasse Y, Schiffmann SN, De Kerchove D'ExaErde A, Qu W, Urade Y, Lazarus M, Huang Z, et al. Projections of nucleus accumbens adenosine A2A receptor neurons in the mouse brain and their implications in mediating sleep-wake regulation. Front Neuroanat 2013; 7(43):1-15; PMID: 24409122; http://dx.doi.org/10.3389/fnana.2013.00043 Front Neuroanat. 2013 Dec 10;7:43. doi: 10.3389/fnana.2013.00043. eCollection 2013. PubMed PMID: 24409122; PubMed Central PMCID: PMC3857888.
- Deisseroth K, Feng G, Majewska AK, Miesenböck G, Ting A, Schnitzer MJ. Next-Generation Optical Technologies for Illuminating Genetically Targeted Brain Circuits. J Neurosci 2006; 26:10380-6; PMID:17035522; http://dx.doi.org/10.1523/JNEUROSCI.3863-06.2006
- Pastrana E. Optogenetics: controlling cell function with light. Nat Meth 2011; 8:24-5; http://dx.doi.org/10.1038/nmeth.f.323
- Adamantidis AR, Zhang F, Aravanis AM, Deisseroth K, de Lecea L. Neural substrates of awakening probed with optogenetic control of hypocretin neurons. Nature 2007; 450:420-4; PMID:17943086; http://dx.doi.org/10.1038/nature06310
- Tsunematsu T, Kilduff TS, Boyden ES, Takahashi S, Tominaga M, Yamanaka A. Acute Optogenetic Silencing of Orexin/Hypocretin Neurons Induces Slow-Wave Sleep in Mice. J Neurosci 2011; 31:10529-39; PMID:21775598; http://dx.doi.org/10.1523/JNEUROSCI.0784-11.2011
- Williams RH, Chee MJS, Kroeger D, Ferrari LL, Maratos-Flier E, Scammell TE, Arrigoni E. Optogenetic-Mediated Release of Histamine Reveals Distal and Autoregulatory Mechanisms for Controlling Arousal. J Neurosci 2014; 34:6023-9; PMID:24760861; http://dx.doi.org/10.1523/JNEUROSCI.4838-13.2014
- Anaclet C, Ferrari L, Arrigoni E, Bass CE, Saper CB, Lu J, Fuller PM. The GABAergic parafacial zone is a medullary slow wave sleep-promoting center. Nat Neurosci 2014; 17:1217-24; PMID:25129078; http://dx.doi.org/10.1038/nn.3789
- Zamir N, Skofitsch G, Bannon MJ, Jacobowitz DM. Melanin-concentrating hormone: unique peptide neuronal system in the rat brain and pituitary gland. Proc Natl Acad Sci USA 1986; 83:1528-31; PMID:3513180; http://dx.doi.org/10.1073/pnas.83.5.1528
- Jego S, Glasgow SD, Herrera CG, Ekstrand M, Reed SJ, Boyce R, Friedman J, Burdakov D, Adamantidis AR. Optogenetic identification of a rapid eye movement sleep modulatory circuit in the hypothalamus. Nat Neurosci 2013; 16:1637-43; PMID:24056699; http://dx.doi.org/10.1038/nn.3522
- Konadhode RR, Pelluru D, Blanco-Centurion C, Zayachkivsky A, Liu M, Uhde T, Glen WB, van den Pol AN, Mulholland PJ, Shiromani PJ. Optogenetic Stimulation of MCH Neurons Increases Sleep. J Neurosci 2013; 33:10257-63; PMID:23785141; http://dx.doi.org/10.1523/JNEUROSCI.1225-13.2013
- Tsunematsu T, Ueno T, Tabuchi S, Inutsuka A, Tanaka KF, Hasuwa H, Kilduff TS, Terao A, Yamanaka A. Optogenetic Manipulation of Activity and Temporally Controlled Cell-Specific Ablation Reveal a Role for MCH Neurons in Sleep/Wake Regulation. J Neurosci 2014; 34:6896-909; PMID:24828644; http://dx.doi.org/10.1523/JNEUROSCI.5344-13.2014
- Sprengel R, Hasan MT. Tetracycline-controlled genetic switches. Handb Exp Pharmacol 2007; 178:49-72; PMID:17203651; http://dx.doi.org/10.1007/978-3-540-35109-2_3
- Tanaka KF, Matsui K, Sasaki T, Sano H, Sugio S, Fan K, Hen R, Nakai J, Yanagawa Y, Hasuwa H, et al. Expanding the repertoire of optogenetically targeted cells with an enhanced gene expression system. Cell Rep 2012; 2:397-406; PMID:22854021; http://dx.doi.org/10.1016/j.celrep.2012.06.011
- Tsunematsu T, Tabuchi S, Tanaka KF, Boyden ES, Tominaga M, Yamanaka A. Long-lasting silencing of orexin/hypocretin neurons using archaerhodopsin induces slow-wave sleep in mice. Behav Brain Res 2013; 255:64-74; PMID:23707248; http://dx.doi.org/10.1016/j.bbr.2013.05.021
- Lerchner W, Xiao C, Nashmi R, Slimko EM, van Trigt L, Lester HA, Anderson David J. Reversible Silencing of Neuronal Excitability in Behaving Mice by a Genetically Targeted, Ivermectin-Gated Cl− Channel. Neuron 2007; 54:35-49; PMID:17408576; http://dx.doi.org/10.1016/j.neuron.2007.02.030
- Oishi Y, Williams RH, Agostinelli L, Arrigoni E, Fuller PM, Mochizuki T, Saper CB, Scammell TE. Role of the Medial Prefrontal Cortex in Cataplexy. J Neurosci 2013; 33:9743-51; PMID:23739971; http://dx.doi.org/10.1523/JNEUROSCI.0499-13.2013
- Rogan SC, Roth BL. Remote Control of Neuronal Signaling. Pharmacol Rev 2011; 63:291-315; PMID:21415127; http://dx.doi.org/10.1124/pr.110.003020
- Dong S, Allen JA, Farrell M, Roth BL. A chemical-genetic approach for precise spatio-temporal control of cellular signaling. Mol Bio Systems 2010; 6:1376-80
- Alexander GM, Rogan SC, Abbas AI, Armbruster BN, Pei Y, Allen JA, Nonneman RJ, Hartmann J, Moy SS, Nicolelis MA, et al. Remote Control of Neuronal Activity in Transgenic Mice Expressing Evolved G Protein-Coupled Receptors. Neuron 2009; 63:27-39; PMID:19607790; http://dx.doi.org/10.1016/j.neuron.2009.06.014
- Nawaratne V, Leach K, Suratman N, Loiacono RE, Felder CC, Armbruster BN, Roth BL, Sexton PM, Christopoulos A. New insights into the function of M4 muscarinic acetylcholine receptors gained using a novel allosteric modulator and a DREADD (Designer Receptor Exclusively Activated by a Designer Drug). Mol Pharmacol 2008; 74:1119-31; PMID:18628403; http://dx.doi.org/10.1124/mol.108.049353
- Sasaki K, Suzuki M, Mieda M, Tsujino N, Roth B, Sakurai T. Pharmacogenetic Modulation of Orexin Neurons Alters Sleep/Wakefulness States in Mice. PLoS ONE 2011; 6:e20360; PMID:21647372; http://dx.doi.org/10.1371/journal.pone.0020360
- Inutsuka A, Inui A, Tabuchi S, Tsunematsu T, Lazarus M, Yamanaka A. Concurrent and robust regulation of feeding behaviors and metabolism by orexin neurons. Neuropharmacol 2014; 85:451-60; http://dx.doi.org/10.1016/j.neuropharm.2014.06.015
- Vardy E, Robinson JE, Li C, Olsen Reid HJ, DiBerto Jeffrey F, Giguere Patrick M, Sassano Flori M, Huang X-P, Zhu H, Urban Daniel J, et al. A New DREADD Facilitates the Multiplexed Chemogenetic Interrogation of Behavior. Neuron 2015; 86:936-46; PMID:25937170; http://dx.doi.org/10.1016/j.neuron.2015.03.065
- Sakaguchi M, Kim K, Yu LMY, Hashikawa Y, Sekine Y, Okumura Y, Kawano M, Hayashi M, Kumar D, Boyden ES, et al. Inhibiting the Activity of CA1 Hippocampal Neurons Prevents the Recall of Contextual Fear Memory in Inducible ArchT Transgenic Mice. PLoS ONE 2015; 10:e0130163; PMID:26075894; http://dx.doi.org/10.1371/journal.pone.0130163
- Goto A, Nakahara I, Yamaguchi T, Kamioka Y, Sumiyama K, Matsuda M, Nakanishi S, Funabiki K. Circuit-dependent striatal PKA and ERK signaling underlies rapid behavioral shift in mating reaction of male mice. Proc Natl Acad Sci USA 2015; 112:6718-23; PMID:25964359; http://dx.doi.org/10.1073/pnas.1507121112
- Sweeney ST, Broadie K, Keane J, Niemann H, O'Kane CJ. Targeted expression of tetanus toxin light chain in Drosophila specifically eliminates synaptic transmission and causes behavioral defects. Neuron 1995; 14:341-51; PMID:7857643; http://dx.doi.org/10.1016/0896-6273(95)90290-2
- Hikida T, Kimura K, Wada N, Funabiki K, Nakanishi S. Distinct Roles of Synaptic Transmission in Direct and Indirect Striatal Pathways to Reward and Aversive Behavior. Neuron 2010; 66:896-907; PMID:20620875; http://dx.doi.org/10.1016/j.neuron.2010.05.011
- Garner AR, Rowland DC, Hwang SY, Baumgaertel K, Roth BL, Kentros C, Mayford M. Generation of a Synthetic Memory Trace. Science 2012; 335:1513-6; PMID:22442487; http://dx.doi.org/10.1126/science.1214985
- Ramirez S, Liu X, Lin P-A, Suh J, Pignatelli M, Redondo RL, Ryan TJ, Tonegawa S. Creating a False Memory in the Hippocampus. Science 2013; 341:387-91; PMID:23888038; http://dx.doi.org/10.1126/science.1239073
- Liu X, Ramirez S, Pang PT, Puryear CB, Govindarajan A, Deisseroth K, Tonegawa S. Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature 2012; 484:381-5; PMID:22441246; http://dx.doi.org/10.1038/484410a
- Zhang Z, Ferretti V, Guntan I, Moro A, Steinberg EA, Ye Z, Zecharia AY, Yu X, Vyssotski AL, Brickley SG, et al. Neuronal ensembles sufficient for recovery sleep and the sedative actions of ; alpha2 adrenergic agonists. Nat Neurosci 2015; 18:553-61; PMID:25706476; http://dx.doi.org/10.1038/nn.3957
- Callaway EM. A molecular and genetic arsenal for systems neuroscience. Trends Neurosci 2005; 28:196-201; PMID:15808354; http://dx.doi.org/10.1016/j.tins.2005.01.007
- Huang Z-L, Urade Y, Hayaishi O. Prostaglandins and adenosine in the regulation of sleep and wakefulness. Curr Opin Pharmacol 2007; 7:33-8; PMID:17129762; http://dx.doi.org/10.1016/j.coph.2006.09.004
- Huang ZL, Urade Y, Hayaishi O. The role of adenosine in the regulation of sleep. Curr Top Med Chem 2011; 11:1047-57; PMID:21401496; http://dx.doi.org/10.2174/156802611795347654
- Lazarus M, Huang Z-L, Lu J, Urade Y, Chen J-F. How do the basal ganglia regulate sleep–wake behavior? Trends Neurosci 2012; 35:723-32; PMID:22858523; http://dx.doi.org/10.1016/j.tins.2012.07.001
- Lazarus M, Chen J-F, Urade Y, Huang Z-L. Role of the basal ganglia in the control of sleep and wakefulness. Curr Opin Neurobiol 2013; 23:780-5; PMID:23465424; http://dx.doi.org/10.1016/j.conb.2013.02.001
- Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, Berthold P, Ollig D, Hegemann P, Bamberg E. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci USA 2003; 100:13940-5; PMID:14615590; http://dx.doi.org/10.1073/pnas.1936192100
- Bamann C, Kirsch T, Nagel G, Bamberg E. Spectral Characteristics of the Photocycle of Channelrhodopsin-2 and Its Implication for Channel Function. J Mol Biol 2008; 375:686-94; PMID:18037436; http://dx.doi.org/10.1016/j.jmb.2007.10.072
