Abstract
Homeodomain-interacting protein kinase 2 (HIPK2) is a serine/threonine protein kinase that participates in the regulation of diverse cellular activities as a transcriptional cofactor and signal transducer. HIPK2 senses various signaling cues that in turn phosphorylate downstream substrates to coordinate developmental processes, cell cycle regulation, cell proliferation, differentiation, and the DNA damage response. HIPK2 functions are affected by its catalytic activity, stability, and subcellular localization, which in turn are dynamically regulated by diverse post-translational modifications such as polyubiquitination, SUMOylation, phosphorylation, and acetylation. HIPK2 is not modified with small molecules and/or peptides individually or independently, but in a combinatorial manner that is referred to as the “HIPK2 modification code.” HIPK2 integrates various signaling cues and senses different doses of DNA damage and ROS stimuli, which are reflected by unique patterns of HIPK2 modification. Hence, the HIPK2 modification code differentially contributes to cellular homeostasis and determination of cell fate depending on cellular context.
Abbreviations
| ADR | = | adriamycin |
| AMPKα | = | adenosine monophosphate-activated kinase α |
| ATF | = | activating transcription factor |
| ATM | = | ataxia-telangiectasia mutated |
| ATR | = | ATM- and RAD3-related |
| CREB | = | cAMP response element binding protein |
| DSB | = | double strand break |
| HDAC | = | histone deacetylase |
| HIPK2 | = | homeodomain-interacting protein kinase 2 |
| IR | = | ionizing radiation |
| Mdm2 | = | mouse double minute 2 |
| ROS | = | reactive oxygen species |
| Siah | = | seven in absentia homolog |
| SUMO | = | small ubiquitin-like modifier |
| PML-NB | = | promyelocytic leukemia nuclear body |
| PTM | = | post-translational modification |
| WIP1 | = | wild-type p53-inducible phosphatase 1 |
| WSB-1 | = | WD40 domain and suppressor of cytokine signaling (SOCS) box protein-1 |
Introduction
Our understanding of protein post-translational modification (PTM) has come a long way over the past decades since protein phosphorylation was first recognized as a key regulatory mechanism of diverse signaling pathways. Advances in biochemical techniques including mass spectrometry have enabled researchers to map the sites of modification in physiological conditions as well as in vitro. Protein functions are regulated by differential modifications in response to diverse intrinsic or extrinsic cellular stimuli. Protein modification by small molecules (such as phosphorylation, acetylation, methylation) and by peptides (such as ubiquitination, SUMOylation, ISGylation) occurs dynamically depending on cellular context, even in response to different levels of the same stimulus. Therefore, analysis of the PTM dynamics of key regulatory proteins in diverse signaling pathways is pivotal to understanding the real-time regulation of cellular activity and signal networking in a spatiotemporal manner.
Homeodomain-interacting protein kinase 2 (HIPK2) is a member of a nuclear serine/threonine kinase family containing 4 proteins (HIPK1-HIPK4).Citation1-3 HIPKs were first identified as Nkx1.2-interacting proteins in yeast 2-hybrid screening. HIPKs contain a conserved protein kinase domain separated from a domain that interacts with homeoproteins, and are therefore termed homeodomain-interacting protein kinases.Citation3 Hipk1 or Hipk2 single knockout mice are grossly normal as a result of functional redundancy between HIPK1 and HIPK2.Citation4,5 However, Hipk1/Hipk2 double knockout mice are embryonic lethal and exhibit defective neural tube closure in response to morphogenetic signals during mouse development and defects in differentiation of the haematopoietic cell lineage, and defective vasculogenesis and angiogenesis.Citation6-8 Loss of HIPK2 alone was subsequently reported to result in defects in fetal liver erythropoiesis and progressive loss of enteric neurons during postnatal mouse development.Citation9,10 In addition, several reports suggest a role of HIPK2 in cellular differentiation. HIPK2 is involved in the differentiation of myoblasts and angiogenesis by suppressing MEF2C-dependent transcriptional activation of tissue-specific target genes.Citation8,11 During early neuronal development HIPK2 and phospho-specific prolyl-peptidyl cis/trans isomerase 1 (Pin1) are recruited to the Groucho/TLE1:Hes1 repressor complex where HIPK2 phosphorylates Groucho/TLE1 to promote cortical neurogenesis by suppressing Groucho/TLE-mediated inhibition of neuronal differentiation.Citation12 From HIPK2 knockout studies it is evident that HIPK2 participates in diverse developmental processes and the differentiation of multiple tissue subsets, including neurogenesis, myogenesis, angiogenesis, fat development, and hematopoiesis.Citation11-13
A representative phosphorylation target of HIPK2 is p53. HIPK2 phosphorylates p53 at the Ser46 residue, which is crucial in p53-mediated induction of apoptosis.Citation14,15 To date, more than 20 proteins have been identified as HIPK2 phosphorylation targets. HIPK2 participates in the coordination of diverse developmental signaling pathways including TGF-β, Notch, Wnt, Hedgehog, and Hippo signaling.Citation8,9,Citation16-21 Most studies on HIPK2 phosphorylation of downstream effectors and HIPK2-mediated regulation of target genes at a transcriptional level were interpreted by the simple concept of turning HIPK2 on or off at a switchboard. Recent studies, however, indicate that HIPK2 is an integrating sensor for diverse cellular signaling events such as genotoxic stresses, hypoxia, and reactive oxygen species (ROS). Integration of different doses of signaling cues and damage stimuli is reflected in diverse post-translational modifications, stability, catalytic activity of HIPK2, and the consequent determination of cell fates (). In this review we will focus on the determination of cell fate—cell death or survival—by differential states of diverse HIPK2 post-translational modifications (termed the “HIPK2 modification code”), in particular as determined by different levels of ROS () or DNA damage stimuli ().
Figure 1. Regulation of HIPK2 depending on the level of DNA damage. HIPK2 regulation was initially understood simply as turning ON or OFF the activity of HIPK2 and phosphorylation of downstream targets (left panel). However, recent studies suggest that HIPK2 responds differentially to different dosages of environmental stimuli and by integrating different signaling cues (right panel), and thus determines the fate of cell death or survival depending on cellular context and HIPK2 modification patterns. Blue circles, yellow rectangles, and green triangles indicate phosphorylation, SUMOylation, and acetylation of HIPK2, respectively.
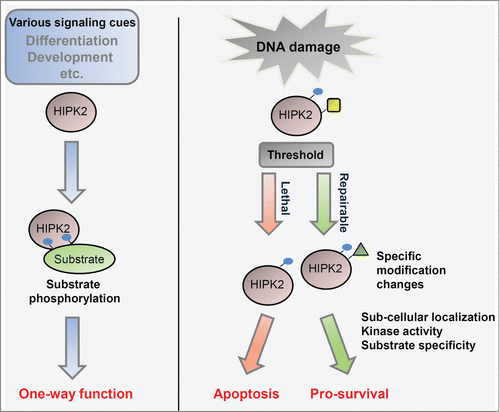
Figure 2. Regulation of cell fate by the HIPK2 modification code depending on different doses of reactive oxygen species. Under normal physiological conditions and basal reactive oxygen species (ROS) levels, HIPK2 is SUMOylated and associated with HDAC3, which maintains HIPK2 in a deacetylated state. In response to an endurable dose of ROS, HIPK2 SUMOylation is replaced by PCAF-mediated HIPK2 acetylation at multiple sites (10K), promoting cell survival. In the presence of a high dose of ROS that is above the threshold for apoptosis induction, HIPK2 acetylation is blocked and unmodified HIPK2 elicits p53-dependent and p53-independent apoptosis. Green triangles and yellow rectangles indicate acetylation and SUMOylation, respectively. HDAC, histone deacetylase; PCAF, P300/CBP-associated factor; PML, promyelocytic leukemia.
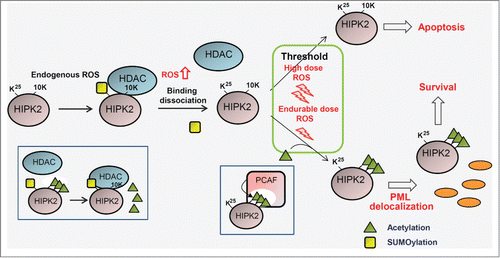
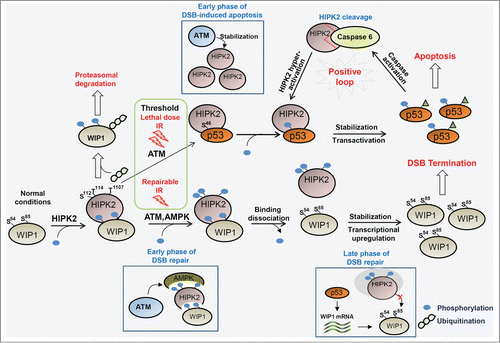
Figure 3. Cell fate is differentially determined by HIPK2 modification and phosphorylation of downstream target proteins in response to ionizing radiation. Under normal growth conditions, WIP1 is constitutively degraded by HIPK2-mediated phosphorylation, which is crucial to maintain an environment favorable for rapid induction of the DNA damage response in response to potential DNA damage. In response to endurable ionizing radiation (IR), DSB signaling is properly activated and DNA repair is completed. WIP1 is then stabilized by escape from HIPK2-mediated degradation and returns cells to a normal state by dephosphorylating ATM and DSB signaling regulators. Upon a high dose of IR that is above the threshold for apoptosis induction, HIPK2 is stabilized and phosphorylates p53 at Ser46 for p53-mediated induction of apoptosis. Therefore, cell fate is differentially regulated by HIPK2 modification and phosphorylation of downstream target proteins. Blue circles and irregular chains of circles indicate phosphorylation and polyubiquitination of HIPK2, respectively. ATM, ataxia-telangiectasia mutated; AMPK, adenosine monophosphate-activated kinase; DSB, double-strand break; WIP-1, wild-type p53-inducible phosphatase 1.
Role of Hipk2 in Unstressed Conditions
Proteasomal degradation of HIPK2 in unstressed conditions
Under normal growth conditions HIPK2 is maintained at low levels and this HIPK2 stabilization results in restriction of cell growth and proliferation.Citation22-24 Tight regulation of HIPK2 levels is achieved by ubiquitination-dependent proteasomal degradation mediated by several E3 ubiquitin ligases. Three E3 ligases are engaged in the regulation of HIPK2 ubiquitination in unstressed conditions (). WD repeat and SOCS box-containing protein 1 (WSB-1) ubiquitinates the C-terminus of HIPK2 and promotes proteasomal degradation. HIPK2 degradation by WSB-1 is terminated in DNA damage conditions, and autophosphorylation of HIPK2 is markedly increased.Citation25 Phosphorylated HIPK2 dissociates from WSB-1, thus relieving HIPK2 from proteasomal degradation. Consistent with these findings, recent papers have reported that autophosphorylation is essential for the catalytic activity of HIPK2 in unstressed cells and for activation of HIPK2 in DNA damage conditions.Citation26–28 Further studies on the correlation between HIPK2 autophosphorylation and its escape from degradation by WSB-1 would enhance our understanding of HIPK2 regulation. WSB-1 is also involved in hypoxia-driven HIPK2 degradation in combination with Siah2.Citation29
Figure 4. The HIPK2 modification code is generated by the integration of different signaling cues and different doses of signaling stimuli. (A) Under normal growth conditions, HIPK2 is maintained at a low level by E3 ubiquitin ligase-mediated proteasomal degradation and a basal level of SUMOylation. (B) HIPK2 is modified differentially depending on the level of DNA damage. Differential modifications of HIPK2 determine the cell fates of survival or death. The HIPK2 modification code shifts dynamically with changes in the cellular environment. Blue circles, yellow rectangles, green triangles, and irregular chains of circles indicate phosphorylation, SUMOylation, acetylation and polyubiquitination of HIPK2, respectively. (C) Summary of the combinatorial modification pattern of HIPK2 for different levels of DNA damage: unstressed, repairable (low level of damage), and apoptotic conditions (high level of damage). The asterisk indicates HIPK2 cleavage that occurs during myogenic differentiation but not under normal culture conditions.
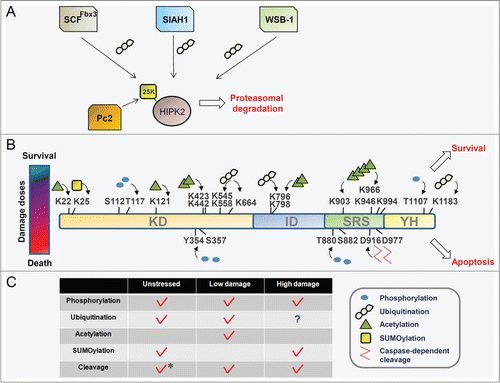
The SCF E3 ligase complex contains Fbx3 together with Cullin1 and Skp1, which were originally isolated as components of the promyelocytic leukemia nuclear body (PML-NB) complex. In unstressed conditions HIPK2, as well as p300, is constitutively degraded by Fbx3-mediated polyubiquitination.Citation30 HIPK2 is degraded outside of the PML-NB because PML-IV prevents HIPK2 destabilization by Fbx3-mediated degradation in the PML-NB. Polyubiquitinated HIPK2 inside the PML-NB is not degraded but instead potentiates p53-mediated transactivation. As recruitment of HIPK2 to PML-NB is a crucial step for the apoptotic response,Citation31 degradation of HIPK2 by Fbx3 might be relieved by SUMOylation-mediated recruitment of HIPK2 to the PML-NB in response to lethal doses of DNA damage stimuli.
Siah1 is another E3 ligase responsible for HIPK2 degradation.Citation32,33 Siah1 degrades HIPK2 constitutively under normal conditions. However, in response to lethal DNA damage ataxia-telangiectasia mutated (ATM) is activated and phosphorylates Siah1 at Ser19. HIPK2 is stabilized by escape from phosphorylated Siah1, and phosphorylates p53 for induction of proapoptotic genes. Downregulation of Siah1 and a reverse correlation between Siah1 and HIPK2 levels were also observed in HIV-expressing transgenic mice and in cells treated with hydrogen peroxide and adriamycin (ADR).Citation18,32 Zyxin indirectly regulates HIPK2 degradation by inhibiting Siah1 activity. Ectopically expressed Zyxin inactivates Siah1 by interfering with Siah1 dimerization and consequently stabilizes HIPK2.Citation34
HIPK2 controls cytokinesis and expression of redox-regulating genes
Although HIPK2 is maintained at low levels under normal growth conditions, it has significant roles in the proper progression of the cell cycle and maintaining cellular homeostasis by preparing cells for potential DNA damage. Reactive oxygen species play versatile roles in cells as a second messenger in signaling pathways.Citation35 At higher concentrations ROS induce cell death, thus preventing the genomic instability caused by ROS-mediated DNA damage.Citation36 HIPK2 functions differentially in response to ROS in a dose-dependent manner (). In unstressed normal growth conditions (physiological low levels of ROS), HIPK2 is predominantly SUMOylated and associated with HDAC3, which maintains HIPK2 in a deacetylated state. Unacetylated HIPK2 functions as a master redox transcriptional regulator that decreases ROS levels and differentially controls the expression of redox-regulating genes such as heme oxygenase 1, peroxiredoxin3, and glutathione S-transferase α1.
Although HIPK2 is associated with cell cycle checkpoint activation in response to DNA damage, it is also involved in cell cycle regulation including cytokinesis.Citation37 During cytokinesis, HIPK2 phosphorylates histone 2B at Ser14, enabling cells to progress through cell cleavage and preventing tetraploidization. Depletion of HIPK2 was also reported to induce p21-dependent cycle arrest.Citation38 Since p21-mediated cell cycle arrest is induced in the absence or presence of HIPK2, both the levels and activity of HIPK2 appear to be important in cell cycle progression.
HIPK2 controls WIP1 homeostasis by phosphorylation-mediated proteasomal degradation
Recent studies revealed another layer of HIPK2 function in the maintenance of cellular homeostasis by preparing cells for potential DNA damage. DNA double-strand breaks (DSB) are induced by ionizing radiation (IR) or other DNA damaging reagents and are repaired by homologous recombination (HR) or nonhomologous end joining (NHEJ) depending on the cell cycle. DSB-driven recruitment of the Mre11-Rad50-Nbs1 (MRN) complex and activation of the apical protein kinase ATM promotes rapid phosphorylation cascades to induce cell cycle arrest and recruitment of repair machinery to the sites of DNA damage.Citation39 DSB signaling is initiated by ATM, which is maintained in an inert state in unstressed conditions. Wild-type p53-inducible phosphatase 1 (WIP1) is a negative regulator that reverses the phosphorylation of ATM and its downstream targets.Citation40,41 WIP1 must be maintained at low levels for the rapid activation of ATM and regulators of DSB signaling. Under normal growth conditions, HIPK2 controls WIP1 homeostasis by phosphorylation-mediated proteasomal degradation (). HIPK2 phosphorylates WIP1 at Ser54 and Ser85, and phosphorylated WIP1 is subject to proteasomal degradation so that WIP1 is maintained at low levels. This silencing of WIP1 function is required for rapid and full activation of DSB signaling regulators in the early phase of the DNA damage response. Depletion of HIPK2 induces WIP1 stabilization, which suppresses DSB signaling and in turn decreases the survival rate of cells exposed to IR. In response to IR, phosphorylation of WIP1 by HIPK2 is gradually decreased and the stabilized WIP1 terminates DSB signaling to allow cells to recover to normal conditions. Interestingly, this mechanism is reminiscent of Siah2 regulation by HIPK2, in which HIPK2 phosphorylates and induces degradation of Siah2 in unstressed conditions and this is reversed by Siah2-mediated HIPK2 degradation in response to hypoxia.Citation42
In summary, under normal growth conditions, HIPK2 is autophosphorylated at a basal level or exists in a hypophosphorylated state. HIPK2 is continuously polyubiquitinated by 3 E3 ubiquitin ligases and subjected to proteasomal degradation to maintain a low expression level. Nonetheless, in normal conditions HIPK2 plays a significant role in the protection of cells from DNA damage. Conjugation of HIPK2 by SUMO-1 antagonizes ROS-triggered HIPK2 acetylation, and HIPK2-mediated WIP1 phosphorylation maintains WIP1 at low levels to ensure a rapid response to ionizing radiation.
Prosurvival Function of HIPK2 in Response to Endurable DNA Damage
HIPK2 induces cell cycle arrest and expression of repair enzymes
Mammalian cells have developed elaborate molecular networks to cope with endurable DNA damage. The DNA damage checkpoint is activated to induce cell cycle arrest and expression of regulatory proteins for DNA repair. HIPK2 is a component of the molecular network that responds to repairable DNA damage. To arrest the cell cycle, p21 expression is induced in multiple ways. HIPK2 induces P300/CBP-associated factor (PCAF)-mediated acetylation of p53 at the Lys320 residue and proteasomal degradation of the p21 transcription repressor ZBTB4,Citation43 both of which contribute to induction of p21 expression. Members of the activating transcription factor/cAMP response element binding protein (ATF/CREB) family are phosphorylation targets of HIPK2 that function to improve cell survival. HIPK2 induces expression of the BDNF growth factor by phosphorylating CREB at Ser271 in response to etoposide treatment. Under conditions of repairable DNA damage, an antioxidant environment is also regulated by HIPK2, which phosphorylates ATF1 at Ser198. Phosphorylated ATF1 potentiates the expression of a series of antioxidant detoxification enzymes such as NADPH quinone oxidoreductase 1, glutathione S transferase, and heme oxygenase 1. HIPK2 can function at the transcriptional level as a transcriptional corepressor and coactivator in a context-dependent manner.Citation44,45 HIPK2-mediated expression of the repair enzyme p53R2 also contributes to DNA damage repair and the maintenance of genome integrity.Citation46
HIPK2 modification under conditions of repairable DNA damage
AMP-activated protein kinase (AMPK) is a versatile protein kinase responsible for numerous signaling pathways, in particular those involved in glucose sensing and autophagy induction.Citation47,48 Upon the generation of DSBs caused by repairable doses of IR, ATM phosphorylates AMPKα, which in turn phosphorylates HIPK2 at 3 sites (T112, S114, and T1107). Phosphorylated HIPK2 dissociates from WIP1 (), preventing HIPK2-mediated phosphorylation and degradation of WIP1 and thus leading to WIP1 stabilization and termination of DSB repair signaling. In the late stage of the DSB response, transcriptional induction of WIP1 by p53 also assists in increasing the levels of WIP1.Citation49,50
HIPK2 also shows altered modification patterns in response to endurable ROS levels (). At a physiological level of ROS under normal growth conditions, SUMOylated HIPK2 recruits HDAC3 to maintain HIPK2 in a deacetylated state. In response to elevated ROS, however, HIPK2 is deSUMOylated and acetylated at 10 lysine residues by CBP acetyl-transferase. Alteration of the HIPK2 modification pattern from SUMOylation to acetylation results in a shift in HIPK2 localization from nuclear speckles to the nucleoplasm and the cytoplasm and release of HIPK2-mediated transcriptional repression of several redox-regulating genes. Proper acetylation of HIPK2 at endurable ROS doses is essential for tolerance against ROS-induced cell death. HIPK2 SUMOylation performs additional roles in response to genotoxic stress. HIPK2 phosphorylates the Pc2 E3 SUMO ligase and enhances its enzymatic activity. Pc2 in turn SUMOylates HIPK2, which then participates in the transcriptional repression of proapoptotic genes such as bax to inhibit apoptosis and promote cell survival.Citation51
HIPK2 regulation by the p53-Mdm2 axis
At sublethal doses of ADR, Mdm2-mediated ubiquitination of HIPK2 provides an additional layer of HIPK2 modification.Citation52 Mdm2 is well known as an E3 ligase responsible for p53 degradation and as a transcriptional target of p53.Citation53 The p53 and Mdm2 regulatory loop affects numerous cellular events including tumorigenesis, the DNA damage response, and cell cycle regulation. Since HIPK2 and Mdm2 induce degradation of each other in a reciprocal manner depending on cellular context and are regulators of p53 acting in opposite directions, the mechanism underlying the mutual regulation between HIPK2 and Mdm2 is not simple. At endurable doses of ADR, the p53-Mdm2 equilibrium is shifted toward Mdm2, which inhibits p53-mediated apoptosis and also downregulates HIPK2 through Mdm2-mediated ubiquitination on the HIPK2 Lys1182 residue. In this case, Mdm2 also disrupts the Axin–p53 interaction by competitive binding to p53 independent of its E3 ligase activity.Citation54 Artificial modulation of Mdm2 levels through treatment with RITA and nutlin-1 results in alterations of HIPK2 stability, p53 phosphorylation, and the apoptotic efficiency of tumor cells.Citation55 Consequently, dual regulation of p53 and HIPK2 by Mdm2 in response to repairable doses of DNA damaging agents allows damaged cells to protect themselves from apoptosis and to accelerate a repair program.
Role of HIPK2 in the Apoptotic Response
p53-dependent and -independent induction of apoptosis by HIPK2
Apoptosis must be tightly controlled because deregulated apoptosis can cause developmental defects and diverse human diseases. Alteration of HIPK2 modifications in response to apoptotic stimuli leads to changes in both the amount and biochemical properties of HIPK2. HIPK2 is stabilized by escape from E3 ligase-mediated proteasomal degradation. During HIPK2 stabilization, HIPK2 autophosphorylation and phosphorylation of E3 ubiquitin ligases by ATM allow HIPK2 to dissociate from E3 ubiquitin ligases. Upon DNA damage, HIPK2 phosphorylation is induced by protein kinases such as AMPK, Src, and TAK1.Citation8,16,56,57 HIPK2 induces apoptosis in both a p53-dependent and p53-independent manner. The HIPK2–p53 axis constitutes the central regulatory axis for p53-mediated induction of apoptosis. p53 protein that is phosphorylated at Ser46 is specifically recruited to the promoters of proapoptotic genes to induce apoptosis at the transcriptional level. Several negative and positive regulators of the HIPK2–p53 axis have been shown to modulate and fine-tune HIPK2–p53-dependent apoptosis.
It is well established that Ser 46 phosphorylation and stabilization of p53 in the PML-NB are the predominant roles of HIPK2 in apoptotic conditions.Citation14,15,31,58 A macromolecular complex including Axin and Daxx is involved in this mechanism.Citation36,59 Axin is an adaptor for formation of the Axin–HIPK2–p53 complex that is important for HIPK2-mediated p53 phosphorylation in the PML-NB. The formation of this complex is inhibited by the negative regulator Pirh2 in sublethal damage conditions and potentiated by the positive regulator Tip60 at lethal levels of DNA damage. At sublethal conditions, Pirh2 competes with HIPK2 for Axin binding and inhibits p53 Ser46 phosphorylation. At lethal doses of DNA damage, however, Tip60-mediated inhibition of the Pirh2-Axin interaction promotes HIPK2–Axin binding and induces p53 Ser46 phosphorylation in an ATM/ATR-dependent manner. Truncation of the Axin HIPK2-interacting domain impairs HIPK2-mediated p53 phosphorylation, indicating that Axin and HIPK2 are critical determinants of cell fate depending on the severity of genotoxic stresses. Daxx is also important for HIPK2-mediated phosphorylation of p53 by serving as a bridge between p53 and Axin.Citation36 In addition, the integrity of PML-NB is also regulated by HIPK2-dependent PML phosphorylation. During early stages of DNA damage, HIPK2 phosphorylates PML at Ser8 and Ser38 to induce PML stabilization and SUMOylation for the induction of apoptosis.Citation60
In response to lethal doses of genotoxic stresses, HIPK2 is stabilized by escape from proteasomal degradation and induces apoptosis through dual regulation of p53 and its negative regulator Mdm2. HIPK2 phosphorylates Mdm2, resulting in its nuclear export and degradation. The accumulated HIPK2 also phosphorylates p53 at the Ser46 residue. The simultaneous regulation of p53 and Mdm2 by HIPK2 leads to efficient induction of apoptosis.Citation52,Citation61–64
The various functions of p53 under DNA damage conditions are not limited to the Ser46 phosphoprotein. Acetylation of p53 at Lys382 is also important for cell cycle arrest through transcriptional activation of p21Citation65 and recruitment of p53 to the promoters of pro-apoptotic genes such as Noxa and p53AIP.Citation66 HIPK2 affects p53 acetylation by modulating p300/CBPCitation6,14 and PCAFCitation67, which acetylate p53 at different sites. HIPK2 also suppresses expression of Nox1, a catalytic subunit of NADPH oxidase, and consequently inhibits SIRT1 for p53 deacetylation,Citation68 indicating that HIPK2 potentiates p53 acetylation by activation of p53 acetyl-transferase and by inhibition of the p53 deacetylating enzyme SIRT1. In addition, HIPK2 also participates in termination of the p53 response to avoid prolonged and exaggerated p53 activity by indirectly modulating p53 acetylation. HIPK2 restricts p53 acetylation by phosphorylating Siah2 at 5 serine residues, which potentiates the E3 ligase activity of Siah2 toward lysine acetyl-transferases, including p300/CBP, PCAF, and Tip60.Citation69 Several reports suggest that HIPK2-mediated phosphorylation and acetylation of p53 cooperatively and/or differentially contribute to the DNA damage response depending on the character of the DNA damaging reagents.Citation66,67
Regulation of apoptosis by HIPK2 is not only dependent on p53. Expression of proapoptotic genes is suppressed by the transcriptional corepressor C-terminal binding protein (CtBP). Under DNA damage conditions, HIPK2 phosphorylates the anti-apoptotic CtBP protein.Citation70 Phosphorylated CtBP is degraded in a proteasome-dependent manner which in turn relieves CtBP-mediated downregulation of several proapoptotic genes such as PERP, p21, and Noxa.Citation71 Therefore, HIPK2 induces apoptosis in p53 null cells through CtBP degradation in response to apoptotic stimuli. Upon TGF-β treatment, HIPK phosphorylates Daxx and releases it from PML-NB. The released Daxx translocates to the cytoplasm and activates JNK, which plays an important role in TGF-β–induced apoptosis.Citation72
HIPK2 cleavage under DNA damage conditions and cellular differentiation
Cleavage of the autoinhibitory domain of HIPK2 adds yet another layer of HIPK2 modification in response to lethal damage.Citation73 Caspase-6 is involved in the irreversible cleavage of HIPK2 at Asp916 and Asp977. Caspase-mediated cleavage and the consequent removal of the HIPK2 autoinhibitory domain results in a catalytically hyperactive HIPK2 protein that highly phosphorylates p53. As caspase-6 is a transcriptional target of p53,Citation74 positive feedback amplification of the p53-HIPK2 loop increases the sensitivity of the apoptotic response. Caspase-mediated HIPK2 cleavage is also involved in myoblast differentiation as full-length HIPK2 is required for repression of myogenic genes and gradual induction of HIPK2 cleavage results in induction of myogenic gene expression as a result of defective corepressor function of the truncated HIPK2 protein ().Citation11
Several reports have proposed the concept that HIPK2 modifications affect HIPK2 function in a combinational and complicated manner, rather than independently or individually, and thus collectively determine cell fate after differential doses of DNA damage.Citation56,75 In contrast to HIPK2-mediated induction of apoptosis under lethal DNA damage conditions, endurable levels of DNA damage alter HIPK2 kinetics to a prosurvival function through a combination of post-translational modifications.
HIPK2 Autophosphorylation and Transphosphorylation by Other Protein Kinases
Phosphorylations of HIPK2 are among the major modifications required for induction of apoptosis in lethal DNA damage conditions. It was recently reported that HIPK2 is regulated by both autophosphorylation and transphosphorylation by other protein kinases under normal and DNA damage conditions. HIPK2 phosphorylation at Thr880 and Ser882 is induced in response to genotoxic stress and is crucial for p53 Ser46 phosphorylation and induction of apoptosis.Citation26 Treatment of cells with lethal doses of ADR induces oligomerization and autophosphorylation of HIPK2. HIPK2 phosphorylated at T880/S882 is recognized by Pin1, which stabilizes HIPK2 through the induction of isomerization and conformational changes in HIPK2 and dissociation of Siah1. At later stages of the DNA damage response HIPK2 phosphorylates p53 at the Ser46 residue, which acts as another target of Pin1 to synergistically activate the apoptotic process.
Although the HIPK family shows amino acid similarity with the dual-specificity tyrosine phosphorylation-regulated kinase (DYRK) family,Citation76 HIPK2 differs in the mode of action in its activation loop. Under normal conditions, cis-phosphorylation of Tyr354 in the activation loop is critical for HIPK2 kinase activity and subcellular localization.Citation27,28 Substitution of the Tyr354 residue to phenylalanine results in cytoplasmic localization of HIPK2. Tyr354 is a target of TGF-β-induced TAK1Citation8 and Src kinase, pivotal enzymes for cell survival and development.Citation57 Phosphorylation of Tyr354, as well as other tyrosine residues, by Src results in inactivation of HIPK2 through translocation from the nucleus to the cytosol. However, phosphorylation of Tyr354 by TGF-β-induced TAK1 is important in the transcriptional suppression of several potent angiogenic genes such as Mmp10 and Vegf. These discrepancies in the outcome of Tyr354 phosphorylation induced by different signaling cues might be explained by differential phosphorylation of HIPK2 at other sites in addition to Tyr354. In addition, phosphorylation of HIPK2 by AMPK in response to repairable doses of IR enables dissociation of WIP1 from HIPK2 to terminate DSB signaling after completion of DNA repair.Citation56 Therefore, HIPK2 phosphorylation by cis-autophosphorylation or transphosphorylation at different sites may differentially affect HIPK2 activity, substrate recognition, and the function of HIPK2 in the determination of cell fate.
It should be noted that identification of sites of post-translational protein modification is usually conducted with overexpressed affinity-purified protein because of limited quantities of endogenous protein. However, depending on the experimental conditions overexpressed HIPK2 does not necessarily recapitulate endogenous HIPK2, especially regarding localization and post-translational modifications, because HIPK2 levels are critical for its function and post-translational modification. This might explain the discrepancy in the patterns of HIPK2 modification and tentative HIPK2 functions proposed by different researchers. Since HIPK2 is dynamically regulated at the protein level and by post-translational modification in response to various signaling cues, HIPK2 function should ideally be addressed under physiological conditions.
Concluding Remarks and Future Perspectives
Cellular responses to variable amounts of DNA damage are crucial to protect organisms from genomic instability.Citation77 The balance between antagonistic enzymatic functions, such as protein kinases versus phosphatases or acetyl-transferases vs. deacetylating enzymes, is shifted to cope with damage stimuli and either protect cells from death or promote it. Such signal sensors integrate the severity of DNA damage or recognize the cellular threshold of the DNA damage response and determine whether the cell will die or live. HIPK2 is an integrator of several signaling pathways, especially the pathways involved in DNA damage caused by ROS or IR.Citation22,78 Recent studies reveal that integration of DNA damage stimuli is mediated by various reversible modifications on HIPK2 (). However, findings that support the existence of a “HIPK2 modification code” () raise further questions. The first concerns the regulatory factor responsible for the shift in the HIPK2 modification code. HIPK2 autophosphorylation and transphosphorylations by other protein kinases are associated with HIPK2 stability, cellular localization, and corepressor activity. Reversible conjugation of HIPK2 to the SUMO moiety can be regulated by Pc2,Citation51 SENP1, and SENP2,Citation79,80 and HIPK2 SUMOylation determines HIPK2 acetylation, which is crucial for the protective function of HIPK2 depending on ROS concentration. Therefore, deciphering the molecular networks linking different post-translational modifications will further unveil the HIPK2 modification code and provide greater understanding of the basis for the fine-tuning of HIPK2 as a signaling hub. The second question concerns whether other modifications are involved in HIPK2 regulation. As HIPK2 is frequently regulated by alteration of its cellular localization and activity, it is possible that HIPK2 may be modified by other covalent modifications such as ISGylation, monoubiquitination, or methylation. Intensive studies to obtain answers to these questions will provide valuable clues to understanding the sophisticated functions of HIPK2, and might open up the possibility of using HIPK2 as a crucial marker for cellular states exposed to complicated DNA damage stimuli.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Additional information
Funding
References
- Arai S, Matsushita A, Du K, Yagi K, Okazaki Y, Kurokawa R. Novel homeodomain-interacting protein kinase family member, HIPK4, phosphorylates human p53 at serine 9. FEBS Lett 2007; 581:5649-57; PMID:18022393; http://dx.doi.org/10.1016/j.febslet.2007.11.022
- He Q, Shi J, Sun H, An J, Huang Y, Sheikh MS. Characterization of human homeodomain-interacting protein kinase 4 (HIPK4) as a unique member of the HIPK family. Mol Cell Pharmacol 2010; 2:61-8; PMID:20508833
- Kim YH, Choi CY, Lee SJ, Conti MA, Kim Y. Homeodomain-interacting protein kinases, a novel family of co-repressors for homeodomain transcription factors. J Biol Chem 1998; 273:25875-9; PMID:9748262; http://dx.doi.org/10.1074/jbc.273.40.25875
- Wiggins AK, Wei G, Doxakis E, Wong C, Tang AA, Zang K, Luo EJ, Neve RL, Reichardt LF, Huang EJ. Interaction of Brn3a and HIPK2 mediates transcriptional repression of sensory neuron survival. J Cell Biol 2004; 167:257-67; PMID:15492043; http://dx.doi.org/10.1083/jcb.200406131
- Kondo S, Lu Y, Debbas M, Lin AW, Sarosi I, Itie A, Wakeham A, Tuan J, Saris C, Elliott G, et al. Characterization of cells and gene-targeted mice deficient for the p53-binding kinase homeodomain-interacting protein kinase 1 (HIPK1). Proc Natl Acad Sci U S A 2003; 100:5431-6; PMID:12702766; http://dx.doi.org/10.1073/pnas.0530308100
- Aikawa Y, Nguyen LA, Isono K, Takakura N, Tagata Y, Schmitz ML, Koseki H, Kitabayashi I. Roles of HIPK1 and HIPK2 in AML1- and p300-dependent transcription, hematopoiesis and blood vessel formation. EMBO J 2006; 25:3955-65; PMID:16917507; http://dx.doi.org/10.1038/sj.emboj.7601273
- Isono K, Nemoto K, Li Y, Takada Y, Suzuki R, Katsuki M, Nakagawara A, Koseki H. Overlapping roles for homeodomain-interacting protein kinases hipk1 and hipk2 in the mediation of cell growth in response to morphogenetic and genotoxic signals. Mol Cell Biol 2006; 26:2758-71; PMID:16537918; http://dx.doi.org/10.1128/MCB.26.7.2758-2771.2006
- Shang Y, Doan CN, Arnold TD, Lee S, Tang AA, Reichardt LF, Huang EJ. Transcriptional corepressors HIPK1 and HIPK2 control angiogenesis via TGF-beta-TAK1-dependent mechanism. PLoS Biol 2013; 11:e1001527; PMID:23565059; http://dx.doi.org/10.1371/journal.pbio.1001527
- Chalazonitis A, Tang AA, Shang Y, Pham TD, Hsieh I, Setlik W, Gershon MD, Huang EJ. Homeodomain interacting protein kinase 2 regulates postnatal development of enteric dopaminergic neurons and glia via BMP signaling. J Neurosci: Official J Soc Neurosci 2011; 31:13746-57; PMID:21957238; http://dx.doi.org/10.1523/JNEUROSCI.1078-11.2011
- Hattangadi SM, Burke KA, Lodish HF. Homeodomain-interacting protein kinase 2 plays an important role in normal terminal erythroid differentiation. Blood 2010; 115:4853-61; PMID:20231426; http://dx.doi.org/10.1182/blood-2009-07-235093
- de la Vega L, Hornung J, Kremmer E, Milanovic M, Schmitz ML. Homeodomain-interacting protein kinase 2-dependent repression of myogenic differentiation is relieved by its caspase-mediated cleavage. Nucleic Acids Res 2013; 41:5731-45; PMID:23620283; http://dx.doi.org/10.1093/nar/gkt262
- Ciarapica R, Methot L, Tang Y, Lo R, Dali R, Buscarlet M, Locatelli F, del Sal G, Rota R, Stifani S. Prolyl isomerase Pin1 and protein kinase HIPK2 cooperate to promote cortical neurogenesis by suppressing GrouchoTLE: Hes1-mediated inhibition of neuronal differentiation. Cell Death Differ 2014; 21:321-32; PMID:24270405; http://dx.doi.org/10.1038/cdd.2013.160
- Sjolund J, Pelorosso FG, Quigley DA, DelRosario R, Balmain A. Identification of Hipk2 as an essential regulator of white fat development. Proc Natl Acad Sci U S A 2014; 111:7373-8; PMID:24785298; http://dx.doi.org/10.1073/pnas.1322275111
- Hofmann TG, Moller A, Sirma H, Zentgraf H, Taya Y, Droge W, Will H, Schmitz ML. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat Cell Biol 2002; 4:1-10; PMID:11740489; http://dx.doi.org/10.1038/ncb715
- D’Orazi G, Cecchinelli B, Bruno T, Manni I, Higashimoto Y, Saito S, Gostissa M, Coen S, Marchetti A, Del Sal G, et al. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat Cell Biol 2002; 4:11-9; PMID:11780126; http://dx.doi.org/10.1038/ncb714
- Kanei-Ishii C, Ninomiya-Tsuji J, Tanikawa J, Nomura T, Ishitani T, Kishida S, Kokura K, Kurahashi T, Ichikawa-Iwata E, Kim Y, et al. Wnt-1 signal induces phosphorylation and degradation of c-Myb protein via TAK1, HIPK2, and NLK. Genes Dev 2004; 18:816-29; PMID:15082531; http://dx.doi.org/10.1101/gad.1170604
- Lee W, Andrews BC, Faust M, Walldorf U, Verheyen EM. Hipk is an essential protein that promotes Notch signal transduction in the Drosophila eye by inhibition of the global co-repressor Groucho. Dev Biol 2009; 325:263-72; PMID:19013449; http://dx.doi.org/10.1016/j.ydbio.2008.10.029
- Jin Y, Ratnam K, Chuang PY, Fan Y, Zhong Y, Dai Y, Mazloom AR, Chen EY, D’Agati V, Xiong H, et al. A systems approach identifies HIPK2 as a key regulator of kidney fibrosis. Nat Med 2012; 18:580-8; PMID:22406746; http://dx.doi.org/10.1038/nm.2685
- Swarup S, Verheyen EM. Drosophila homeodomain-interacting protein kinase inhibits the Skp1-Cul1-F-box E3 ligase complex to dually promote Wingless and Hedgehog signaling. Proc Natl Acad Sci U S A 2011; 108:9887-92; PMID:21628596; http://dx.doi.org/10.1073/pnas.1017548108
- Poon CL, Zhang X, Lin JI, Manning SA, Harvey KF. Homeodomain-interacting protein kinase regulates Hippo pathway-dependent tissue growth. Curr Biol: CB 2012; 22:1587-94; PMID:22840515; http://dx.doi.org/10.1016/j.cub.2012.06.075
- Chen J, Verheyen EM. Homeodomain-interacting protein kinase regulates Yorkie activity to promote tissue growth. Curr Biol: CB 2012; 22:1582-6; PMID:22840522; http://dx.doi.org/10.1016/j.cub.2012.06.074
- Sombroek D, Hofmann TG. How cells switch HIPK2 on and off. Cell Death Differ 2009; 16:187-94; PMID:18974774; http://dx.doi.org/10.1038/cdd.2008.154
- Calzado MA, Renner F, Roscic A, Schmitz ML. HIPK2: a versatile switchboard regulating the transcription machinery and cell death. Cell Cycle 2007; 6:139-43; PMID:17245128; http://dx.doi.org/10.4161/cc.6.2.3788
- Puca R, Nardinocchi L, Givol D, D’Orazi G. Regulation of p53 activity by HIPK2: molecular mechanisms and therapeutical implications in human cancer cells. Oncogene 2010; 29:4378-87; PMID:20514025; http://dx.doi.org/10.1038/onc.2010.183
- Choi DW, Seo YM, Kim EA, Sung KS, Ahn JW, Park SJ, Lee SR, Choi CY. Ubiquitination and degradation of homeodomain-interacting protein kinase 2 by WD40 repeatSOCS box protein WSB-1. J Biol Chem 2008; 283:4682-9; PMID:18093972; http://dx.doi.org/10.1074/jbc.M708873200
- Bitomsky N, Conrad E, Moritz C, Polonio-Vallon T, Sombroek D, Schultheiss K, Glas C, Greiner V, Herbel C, Mantovani F, et al. Autophosphorylation and Pin1 binding coordinate DNA damage-induced HIPK2 activation and cell death. Proc Natl Acad Sci U S A 2013; 110:E4203-12; PMID:24145406; http://dx.doi.org/10.1073/pnas.1310001110
- Saul VV, de la Vega L, Milanovic M, Kruger M, Braun T, Fritz-Wolf K, Becker K, Schmitz ML. HIPK2 kinase activity depends on cis-autophosphorylation of its activation loop. J Mol Cell Biol 2013; 5:27-38; PMID:23000554; http://dx.doi.org/10.1093/jmcb/mjs053
- Siepi F, Gatti V, Camerini S, Crescenzi M, Soddu S. HIPK2 catalytic activity and subcellular localization are regulated by activation-loop Y354 autophosphorylation. Biochim Biophys Acta 2013; 1833:1443-53; PMID:23485397; http://dx.doi.org/10.1016/j.bbamcr.2013.02.018
- Tong Y, Li QG, Xing TY, Zhang M, Zhang JJ, Xia Q. HIF1 regulates WSB-1 expression to promote hypoxia-induced chemoresistance in hepatocellular carcinoma cells. FEBS Lett 2013; 587:2530-5; PMID:23792163; http://dx.doi.org/10.1016/j.febslet.2013.06.017
- Shima Y, Shima T, Chiba T, Irimura T, Pandolfi PP, Kitabayashi I. PML activates transcription by protecting HIPK2 and p300 from SCFFbx3-mediated degradation. Mol Cell Biol 2008; 28:7126-38; PMID:18809579; http://dx.doi.org/10.1128/MCB.00897-08
- Möller A SH, Hofmann TG, Rueffer S, Klimczak E, Dröge W, Will H, Schmitz ML. PML is required for homeodomain-interacting protein kinase 2 (HIPK2)-mediated p53 phosphorylation and cell cycle arrest but is dispensable for the formation of HIPK domains. Cancer Res 2003; 1:4310-4; PMID:12907596
- Kim SY, Choi DW, Kim EA, Choi CY. Stabilization of HIPK2 by escape from proteasomal degradation mediated by the E3 ubiquitin ligase Siah1. Cancer Lett 2009; 279:177-84; PMID:19250734; http://dx.doi.org/10.1016/j.canlet.2009.01.036
- Winter M, Sombroek D, Dauth I, Moehlenbrink J, Scheuermann K, Crone J, Hofmann TG. Control of HIPK2 stability by ubiquitin ligase Siah-1 and checkpoint kinases ATM and ATR. Nat Cell Biol 2008; 10:812-24; PMID:18536714; http://dx.doi.org/10.1038/ncb1743
- Crone J, Glas C, Schultheiss K, Moehlenbrink J, Krieghoff-Henning E, Hofmann TG. Zyxin is a critical regulator of the apoptotic HIPK2-p53 signaling axis. Cancer research 2011; 71:2350-9; PMID:21248071; http://dx.doi.org/10.1158/0008-5472.CAN-10-3486
- D’Autreaux B, Toledano MB. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat Rev Mol Cell Biol 2007; 8:813-24; PMID:17848967; http://dx.doi.org/10.1038/nrm2256
- Li Q, Lin S, Wang X, Lian G, Lu Z, Guo H, Ruan K, Wang Y, Ye Z, Han J, et al. Axin determines cell fate by controlling the p53 activation threshold after DNA damage. Nat Cell Biol 2009; 11:1128-34; PMID:19731416; http://dx.doi.org/10.1038/ncb1927
- Rinaldo C, Moncada A, Gradi A, Ciuffini L, D’Eliseo D, Siepi F, Prodosmo A, Giorgi A, Pierantoni GM, Trapasso F, et al. HIPK2 controls cytokinesis and prevents tetraploidization by phosphorylating histone H2B at the midbody. Mol Cell 2012; 47:87-98; PMID:22658722
- Iacovelli S, Ciuffini L, Lazzari C, Bracaglia G, Rinaldo C, Prodosmo A, Bartolazzi A, Sacchi A, Soddu S. HIPK2 is involved in cell proliferation and its suppression promotes growth arrest independently of DNA damage. Cell Proliferat 2009; 42:373-84; PMID:19438900; http://dx.doi.org/10.1111/j.1365-2184.2009.00601.x
- Chapman JR, Taylor MR, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell 2012; 47:497-510; PMID:22920291; http://dx.doi.org/10.1016/j.molcel.2012.07.029
- Shreeram S, Demidov ON, Hee WK, Yamaguchi H, Onishi N, Kek C, Timofeev ON, Dudgeon C, Fornace AJ, Anderson CW, et al. Wip1 phosphatase modulates ATM-dependent signaling pathways. Mol Cell 2006; 23:757-64; PMID:16949371; http://dx.doi.org/10.1016/j.molcel.2006.07.010
- Cha H, Lowe JM, Li H, Lee JS, Belova GI, Bulavin DV, Fornace AJ, Jr. Wip1 directly dephosphorylates gamma-H2AX and attenuates the DNA damage response. Cancer Res 2010; 70:4112-22; PMID:20460517; http://dx.doi.org/10.1158/0008-5472.CAN-09-4244
- Calzado MA, de la Vega L, Moller A, Bowtell DD, Schmitz ML. An inducible autoregulatory loop between HIPK2 and Siah2 at the apex of the hypoxic response. Nat Cell Biol 2009; 11:85-91; PMID:19043406; http://dx.doi.org/10.1038/ncb1816
- Yamada D, Perez-Torrado R, Filion G, Caly M, Jammart B, Devignot V, Sasai N, Ravassard P, Mallet J, Sastre-Garau X, et al. The human protein kinase HIPK2 phosphorylates and downregulates the methyl-binding transcription factor ZBTB4. Oncogene 2009; 28:2535-44; PMID:19448668; http://dx.doi.org/10.1038/onc.2009.109
- Choi CY, Kim YH, Kim YO, Park SJ, Kim EA, Riemenschneider W, Gajewski K, Schulz RA, Kim Y. Phosphorylation by the DHIPK2 protein kinase modulates the corepressor activity of Groucho. J Biol Chem 2005; 280:21427-36; PMID:15802274; http://dx.doi.org/10.1074/jbc.M500496200
- Choi CY, Kim YH, Kwon HJ, Kim Y. The homeodomain protein NK-3 recruits Groucho and a histone deacetylase complex to repress transcription. J Biol Chem 1999; 274:33194-7; PMID:10559189; http://dx.doi.org/10.1074/jbc.274.47.33194
- Nardinocchi L, Puca R, Sacchi A, D’Orazi G. HIPK2 knock-down compromises tumor cell efficiency to repair damaged DNA. Biochem Biophys Res Commun 2007; 361:249-55; PMID:17658469; http://dx.doi.org/10.1016/j.bbrc.2007.07.031
- Inoki K, Kim J, Guan KL. AMPK and mTOR in cellular energy homeostasis and drug targets. Ann Rev Pharmacol Toxicol 2012; 52:381-400; PMID:22017684; http://dx.doi.org/10.1146/annurev-pharmtox-010611-134537
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 2011; 13:132-41; PMID:21258367; http://dx.doi.org/10.1038/ncb2152
- Fiscella M, Zhang H, Fan S, Sakaguchi K, Shen S, Mercer WE, Vande Woude GF, O’Connor PM, Appella E. Wip1, a novel human protein phosphatase that is induced in response to ionizing radiation in a p53-dependent manner. Proc Natl Acad Sci U S A 1997; 94:6048-53; PMID:9177166; http://dx.doi.org/10.1073/pnas.94.12.6048
- Rossi M, Demidov ON, Anderson CW, Appella E, Mazur SJ. Induction of PPM1D following DNA-damaging treatments through a conserved p53 response element coincides with a shift in the use of transcription initiation sites. Nucleic Acids Research 2008; 36:7168-80; PMID:19015127; http://dx.doi.org/10.1093/nar/gkn888
- Roscic A, Moller A, Calzado MA, Renner F, Wimmer VC, Gresko E, Ludi KS, Schmitz ML. Phosphorylation-dependent control of Pc2 SUMO E3 ligase activity by its substrate protein HIPK2. Mol Cell 2006; 24:77-89; PMID:17018294; http://dx.doi.org/10.1016/j.molcel.2006.08.004
- Rinaldo C, Prodosmo A, Mancini F, Iacovelli S, Sacchi A, Moretti F, Soddu S. MDM2-regulated degradation of HIPK2 prevents p53Ser46 phosphorylation and DNA damage-induced apoptosis. Mol Cell 2007; 25:739-50; PMID:17349959; http://dx.doi.org/10.1016/j.molcel.2007.02.008
- Manfredi JJ. The Mdm2-p53 relationship evolves: Mdm2 swings both ways as an oncogene and a tumor suppressor. Genes Dev 2010; 24:1580-9; PMID:20679392
- He Y, Lian G, Lin S, Ye Z, Li Q. MDM2 inhibits axin-induced p53 activation independently of its E3 ligase activity. PloS One 2013; 8:e67529; PMID:23826318; http://dx.doi.org/10.1371/journal.pone.0067529
- Rinaldo C, Prodosmo A, Siepi F, Moncada A, Sacchi A, Selivanova G, Soddu S. HIPK2 regulation by MDM2 determines tumor cell response to the p53-reactivating drugs nutlin-3 and RITA. Cancer Res 2009; 69:6241-8; PMID:19638586; http://dx.doi.org/10.1158/0008-5472.CAN-09-0337
- Choi DW, Na W, Kabir MH, Yi E, Kwon S, Yeom J, Ahn JW, Choi HH, Lee Y, Seo KW, et al. WIP1, a homeostatic regulator of the DNA damage response, is targeted by HIPK2 for phosphorylation and degradation. Mol Cell 2013; 51:374-85; PMID:23871434; http://dx.doi.org/10.1016/j.molcel.2013.06.010
- Polonio-Vallon T, Kirkpatrick J, Krijgsveld J, Hofmann TG. Src kinase modulates the apoptotic p53 pathway by altering HIPK2 localization. Cell Cycle 2014; 13:115-25; PMID:24196445; http://dx.doi.org/10.4161/cc.26857
- Moller A, Sirma H, Hofmann TG, Rueffer S, Klimczak E, Droge W, Will H, Schmitz ML. PML is required for homeodomain-interacting protein kinase 2 (HIPK2)-mediated p53 phosphorylation and cell cycle arrest but is dispensable for the formation of HIPK domains. Cancer Res 2003; 63:4310-4; PMID:12907596
- Li Q, Wang X, Wu X, Rui Y, Liu W, Wang J, Wang X, Liou YC, Ye Z, Lin SC. Daxx cooperates with the AxinHIPK2p53 complex to induce cell death. Cancer Res 2007; 67:66-74; PMID:17210684; http://dx.doi.org/10.1158/0008-5472.CAN-06-1671
- Gresko E, Ritterhoff S, Sevilla-Perez J, Roscic A, Frobius K, Kotevic I, Vichalkovski A, Hess D, Hemmings BA, Schmitz ML. PML tumor suppressor is regulated by HIPK2-mediated phosphorylation in response to DNA damage. Oncogene 2009; 28:698-708; PMID:19015637; http://dx.doi.org/10.1038/onc.2008.420
- Dauth I, Kruger J, Hofmann TG. Homeodomain-interacting protein kinase 2 is the ionizing radiation-activated p53 serine 46 kinase and is regulated by ATM. Cancer Res 2007; 67:2274-9; PMID:17332358; http://dx.doi.org/10.1158/0008-5472.CAN-06-2884
- Di Stefano V, Blandino G, Sacchi A, Soddu S, D’Orazi G. HIPK2 neutralizes MDM2 inhibition rescuing p53 transcriptional activity and apoptotic function. Oncogene 2004; 23:5185-92; PMID:15122315; http://dx.doi.org/10.1038/sj.onc.1207656
- Di Stefano V, Mattiussi M, Sacchi A, D’Orazi G. HIPK2 inhibits both MDM2 gene and protein by, respectively, p53-dependent and independent regulations. FEBS Lett 2005; 579:5473-80; PMID:16212962; http://dx.doi.org/10.1016/j.febslet.2005.09.008
- Zhang XP, Liu F, Wang W. Interplay between Mdm2 and HIPK2 in the DNA damage response. J Roy Soc, Interf Roy Soc 2014; 11; 20140319; PMID:24829283
- Dai C, Gu W. p53 post-translational modification: deregulated in tumorigenesis. Trends Mol Med 2010; 16:528-36; PMID:20932800; http://dx.doi.org/10.1016/j.molmed.2010.09.002
- Puca R, Nardinocchi L, Sacchi A, Rechavi G, Givol D, D’Orazi G. HIPK2 modulates p53 activity towards pro-apoptotic transcription. Mol Cancer 2009; 8:85; PMID:19828042; http://dx.doi.org/10.1186/1476-4598-8-85
- Di Stefano V, Soddu S, Sacchi A, D’Orazi G. HIPK2 contributes to PCAF-mediated p53 acetylation and selective transactivation of p21Waf1 after nonapoptotic DNA damage. Oncogene 2005; 24:5431-42; PMID:15897882; http://dx.doi.org/10.1038/sj.onc.1208717
- Puca R, Nardinocchi L, Starace G, Rechavi G, Sacchi A, Givol D, D’Orazi G. Nox1 is involved in p53 deacetylation and suppression of its transcriptional activity and apoptosis. Free Radical Biol Med 2010; 48:1338-46; PMID:20171273; http://dx.doi.org/10.1016/j.freeradbiomed.2010.02.015
- Grishina I, Debus K, Garcia-Limones C, Schneider C, Shresta A, Garcia C, Calzado MA, Schmitz ML. SIAH-mediated ubiquitination and degradation of acetyl-transferases regulate the p53 response and protein acetylation. Biochim Biophys Acta 2012; 1823:2287-96; PMID:23044042; http://dx.doi.org/10.1016/j.bbamcr.2012.09.011
- Zhang Q, Nottke A, Goodman RH. Homeodomain-interacting protein kinase-2 mediates CtBP phosphorylation and degradation in UV-triggered apoptosis. Proc Natl Acad Sci U S A 2005; 102:2802-7; PMID:15708980; http://dx.doi.org/10.1073/pnas.0409373102
- Chinnadurai G. The transcriptional corepressor CtBP: a foe of multiple tumor suppressors. Cancer Res 2009; 69:731-4; PMID:19155295; http://dx.doi.org/10.1158/0008-5472.CAN-08-3349
- Hofmann TG, Stollberg N, Schmitz ML, Will H. HIPK2 regulates transforming growth factor-beta-induced c-Jun NH(2)-terminal kinase activation and apoptosis in human hepatoma cells. Cancer Res 2003; 63:8271-7; PMID:14678985
- Gresko E, Roscic A, Ritterhoff S, Vichalkovski A, del Sal G, Schmitz ML. Autoregulatory control of the p53 response by caspase-mediated processing of HIPK2. EMBO J 2006; 25:1883-94; PMID:16601678; http://dx.doi.org/10.1038/sj.emboj.7601077
- MacLachlan TK, El-Deiry WS. Apoptotic threshold is lowered by p53 transactivation of caspase-6. Proc Natl Acad Sci U S A 2002; 99:9492-7; PMID:12089322; http://dx.doi.org/10.1073/pnas.132241599
- de la Vega L, Grishina I, Moreno R, Kruger M, Braun T, Schmitz ML. A redox-regulated SUMOacetylation switch of HIPK2 controls the survival threshold to oxidative stress. Mol Cell 2012; 46:472-83; PMID:22503103; http://dx.doi.org/10.1016/j.molcel.2012.03.003
- Schmitz ML, Rodriguez-Gil A, Hornung J. Integration of stress signals by homeodomain interacting protein kinases. Biol Chem 2014; 395:375-86; PMID:24225127
- Zhivotovsky B, Kroemer G. Apoptosis and genomic instability. Nat Rev Mol Cell Biol 2004; 5:752-62; PMID:15340382; http://dx.doi.org/10.1038/nrm1443
- Rinaldo C, Prodosmo A, Siepi F, Soddu S. HIPK2: a multitalented partner for transcription factors in DNA damage response and development. Biochem Cell Biol = Biochim Biol Cell 2007; 85:411-8; PMID:17713576; http://dx.doi.org/10.1139/O07-071
- Kim YH, Sung KS, Lee SJ, Kim YO, Choi CY, Kim Y. Desumoylation of homeodomain-interacting protein kinase 2 (HIPK2) through the cytoplasmic-nuclear shuttling of the SUMO-specific protease SENP1. FEBS Lett 2005; 579:6272-8; PMID:16253240; http://dx.doi.org/10.1016/j.febslet.2005.10.010
- Hofmann TG, Jaffray E, Stollberg N, Hay RT, Will H. Regulation of homeodomain-interacting protein kinase 2 (HIPK2) effector function through dynamic small ubiquitin-related modifier-1 (SUMO-1) modification. J Biol Chem 2005; 280:29224-32; PMID:15958389; http://dx.doi.org/10.1074/jbc.M503921200
