ABSTRACT
Long non-coding RNAs (lncRNAs) exert most of their functions through protein interactions. A better understanding of these interactions will facilitate the development of novel therapeutics. Recently, we described how the lncRNA CCAT2 located at the 8q24 cancer amplicon reprograms cancer metabolism by directly interacting in an allele-specific manner with a protein complex.
Non-coding RNAs (ncRNAs) have emerged over the past decades as master regulators of disease, prevailing over their initial label as “mRNA debris”. Whether it is proliferation, energy metabolism, or metastases, ncRNAs seem to be engaged in every aspect of disease initiation and progression.Citation1,2 Long ncRNAs (lncRNAs), the most abundant class of ncRNAs, have reformed the traditional perception of ncRNAs and the way we study them. We no longer perceive them as “one molecule – one function”, but rather as “one molecule – a diversity of functions”.Citation3 Although our understanding of the mechanisms of action of lncRNAs is in its infancy, accumulating evidence suggests they function through interactions with DNA, proteins, and other RNA molecules (messenger RNAs and microRNAs).Citation3 The lncRNA-protein interactions have been of particular interest to the scientific community as they have advanced the concept of RNA:protein complexes acting as an entity to regulate gene expression. However, the subtle aspects of the lncRNA-protein interactions remain unclear: What determines the specificity of the interaction? Do these interactions mediate the functions of the lncRNA or of the RNA-binding protein (RBP)? How do sequence variations in normal or malignant cells impact the formation of RNA:protein complexes? Finding answers to these and other questions will represent the initiating steps into exploring the druggability of lncRNAs.
Our recently published study “Allele-specific reprograming of cancer metabolism by the long non-coding RNA, CCAT2” contributes to understanding these aspects. To fill the void in our knowledge, we investigated the molecular mechanism by which colon cancer associated transcript 2 (CCAT2) regulates cancer metabolism in an allele-specific manner. As reported by our group in 2013, CCAT2 is transcribed from the 8q24 genomic region harboring the colon cancer-associated SNP rs6983267.Citation4 The distinct associated risk for colorectal cancer (CRC) of the two alleles (G and T) has been documented by several studies;Citation5,6 however, the underlying mechanism of CRC susceptibility is vaguely understood. Starting with the observation that the color of the media of in vitro grown cells changed upon CCAT2 modulation, we evaluated the metabolic phenotypes of cells expressing either the G or T allele. Surprisingly, we discovered that the two alleles rendered discrete metabolic phenotypes both in vitro and in vivo. This represented an exciting finding considering that to date there are no studies reporting the differential regulation of a biological process by distinct alleles of the same gene. One major difference between cells expressing the particular alleles was the manner in which they were metabolizing glutamine. Cells with the CCAT2 G allele seemed to have a more active tricarboxylic acid (TCA) cycle by boosting glutamine metabolism. Naturally, we interrogated the expression of the two alternative splicing isoforms (GAC and KGA) of the rate-limiting enzyme of glutaminolysis, glutaminase (GLS). We found that GAC, the more aggressive and cancer-specific isoform,Citation7 was favored by the CCAT2 G-allele – the cancer risk allele linked to a cancer-specific splicing isoform. We confirmed that the regulatory mechanism was alternative splicing and directed our efforts toward identifying the partnering proteins enabling this process. The cleavage factor I (CFIm) protein complex, known to be involved in precursor mRNA processing, revealed itself as a prime candidate. Using in vitro and biochemical approaches, we demonstrated that CCAT2 interacts with components of the complex—CFIm25 (the 25 kDa subunit encoded by the NUDT21 gene) and CFIm68 (the 68 kDa subunit encoded by the CPSF6 gene)—with allele-specific affinities: the G-allele preferentially bound the CFIm25 subunit and the T-allele had a higher affinity for CFIm68 than the G-allele. This allele-specific affinity appears to be a consequence of changes in the secondary structure of the lncRNA induced by the SNP, although further studies need to be conducted to validate this concept. Nonetheless, our findings provide the first glimpse into how a sequence variation in RNAs can influence RNA-protein interactions and suggest a potential explanation for how disease-associated SNPs function. Furthermore, we observed that CCAT2 bound the pre-mRNA of GLS through short sequences (11–13 nt) found mainly in the introns and that the presence of the target pre-mRNA influences the specificity of the interaction between CCAT2 and the proteins. The formation of a ternary RNA:RNA:protein complex was also insinuated by the atomic force microscopy (AFM) images. Although interaction of a lncRNA with proteins, and indirectly with pre-mRNAs, to regulate alternative splicing of the nascent transcript has been exquisitely described by Guttman’s and Lander’s groups,Citation8 our study not only exposes additional refined characteristics of these types of interactions, but also delivers their biological consequences. Our findings are supported by results obtained from The Cancer Genome Atlas (TCGA) colon cancer dataset and colorectal carcinoma (CRC) patient cohorts. These results reveal that the CCAT2-CFIm-GLS regulatory axis is present in 61% of analyzed cases and that there is a significant association between the rs6983267 SNP genotype and the expression of GAC. However, a recent report introduced CFIm25 as a tumor suppressor in glioblastoma, negatively regulating the expression of GLS via 3´UTR processing.Citation9 Although it may initially seem contradictory to our findings, it has been reported that CCAT2 is not expressed in glioblastoma cell lines. This suggests that interaction with the lncRNA may modulate the function of the protein in a cell-specific way. However, GLS is not solely responsible for the metabolic differences observed between the two alleles; affymetrix HTA 2.0 analysis indicated that the CCAT2:CFIm complex may regulate nascent RNA transcripts converging on the same biological pathway, energy metabolism in this case, much like RNA operons.Citation10
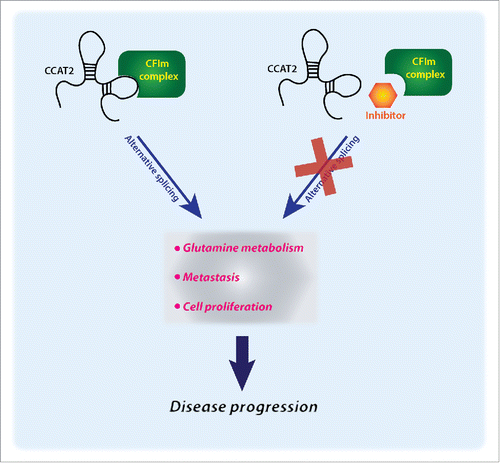
There is growing interest in targeting lncRNAs for therapeutic purposes by disrupting lncRNA-protein interactions. Our study sheds light on the complexity of these interactions and provides a hint to the consequences of reversing them ().
Figure 1. A novel therapeutic concept for long non-coding RNAs: blocking cancer progression through inhibition of the CCAT2:CFIm complex. We hypothesize that by inhibiting the interaction of CCAT2 (colon cancer associated transcript 2) with the CFIm (cleavage factor I) protein complex we could prevent it from exerting its functions and consequently block disease progression.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
Dr Calin is the Alan M. Gewirtz Leukemia & Lymphoma Society Scholar. Work in Dr. Calin’s laboratory is supported in part by the NIH/NCI grants 1UH2TR00943-01 and a R01 CA182905-01, the UT MD Anderson Cancer Center SPORE in Melanoma grant from NCI (P50 CA093459), Aim at Melanoma Foundation and the Miriam and Jim Mulva research funds, the UT MD Anderson Cancer Center Brain SPORE (2P50CA127001), a CLL Global Research Foundation grant, a CLL Moonshot Flagship project, the Laura and John Arnold Foundation, the RGK Foundation, and the Estate of C. G. Johnson, Jr.
References
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genetics 2011; 12:861-74; PMID:22094949; http://dx.doi.org/10.1038/nrg3074
- Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol 2012; 9:703-19; PMID:22664915; http://dx.doi.org/10.4161/rna.20481
- Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol 2013; 14:699-712; PMID:24105322; http://dx.doi.org/10.1038/nrm3679
- Ling H, Spizzo R, Atlasi Y, Nicoloso M, Shimizu M, Redis RS, Nishida N, Gafa R, Song J, Guo Z, et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res 2013; 23:1446-61; PMID:23796952; http://dx.doi.org/10.1101/gr.152942.112
- Haiman CA, Le Marchand L, Yamamato J, Stram DO, Sheng X, Kolonel LN, Wu AH, Reich D, Henderson BE. A common genetic risk factor for colorectal and prostate cancer. Nat Genetics 2007; 39:954-6; PMID:17618282; http://dx.doi.org/10.1038/ng2098
- Tuupanen S, Turunen M, Lehtonen R, Hallikas O, Vanharanta S, Kivioja T, Bjorklund M, Wei G, Yan J, Niittymaki I, et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genetics 2009; 41:885-90; PMID:19561604; http://dx.doi.org/10.1038/ng.406
- van den Heuvel AP, Jing J, Wooster RF, Bachman KE. Analysis of glutamine dependency in non-small cell lung cancer: GLS1 splice variant GAC is essential for cancer cell growth. Cancer Biol Therapy 2012; 13:1185-94; PMID:22892846; http://dx.doi.org/10.4161/cbt.21348
- Engreitz JM, Sirokman K, McDonel P, Shishkin AA, Surka C, Russell P, Grossman SR, Chow AY, Guttman M, Lander ES. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell 2014; 159:188-99; PMID:25259926; http://dx.doi.org/10.1016/j.cell.2014.08.018
- Masamha CP, Xia Z, Yang J, Albrecht TR, Li M, Shyu AB, Li W, Wagner EJ. CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature 2014; 510:412-6; PMID:24814343; http://dx.doi.org/10.1038/nature13261
- Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genetics 2007; 8:533-43; PMID:17572691; http://dx.doi.org/10.1038/nrg2111
