ABSTRACT
The acquisition of resistance to current mitogen activated protein kinase (MAPK) inhibitors in B-Raf proto-oncogene, serine/threonine kinase (BRAF) mutant melanoma is almost inevitable. Our recent findings identify therapy-induced mitochondrial biogenesis (MitoBiogenesis) and aberrant tumor bioenergetics as therapeutic escape mechanisms and offer a rational combinatorial strategy to further improve the efficacy of MAPK inhibitors.
Stage IV melanomas have poor clinical prognosis, although this has improved with new therapies.Citation1,2 Approximately 50% of melanomas harbor a point mutation in the B-Raf proto-oncogene, serine/threonine kinase (BRAF), either V600E or V600K, which results in constitutively active mitogen activated protein kinase (MAPK) signaling.Citation3 Advancements in the development of small-molecule inhibitors targeting mutant BRAF and immune checkpoint blockade therapies targeting immune inhibitory molecules such as cytotoxic T lymphocyte-associated protein 4 (CTLA-4), programmed cell death protein 1 (PD-1), and programmed death-ligand 1 (PD-L1) have provided unprecedented improvements in patient outcomes. These proven therapeutic advances began in 2011, when the BRAF inhibitor vemurafenib was shown to improve both overall and progression-free survival rates for patients with BRAF-mutant melanomas in a phase III clinical trial.Citation4 Subsequently, combined BRAF and mitogen-activated protein kinase kinase (MEK) inhibition, which now is standard of care for treating BRAF-mutant melanomas, has further improved the rate of progression-free survival compared to BRAF inhibition alone.Citation5 Despite the initial clinical efficacy of BRAF-targeted therapies, tumor relapse is almost inevitable due to survival of a small tumor cell population that is dependent on reactivation of the MAPK and phosphatidylinositol-4,5-bisphosphate 3-kinase/protein kinase B (PI3K/AKT) signaling pathways to invade therapy.Citation6
A comprehensive understanding of the molecular mechanisms underlying both intrinsic and acquired drug resistance to MAPK inhibitors (MAPKi) is still being elucidated and will be essential for the development of combinatorial strategies to overcome therapy resistance. Mitochondrial biogenesis (MitoBiogenesis) and tumor metabolism are aberrantly altered in cancer cells under a variety of cellular stress types. The Warburg effect is a shift toward glycolysis that is frequently utilized by many types of tumors, but some melanomas balance their metabolic needs depending on the interplay between microphthalmia-associated transcription factor (MITF), which is a key regulator of MitoBiogenesis, and peroxisome proliferator-activated receptor gamma coactivator 1-α (PPARGC1α).Citation7 Melanoma cells with high expression of MITF and PPARGC1α exhibit increased oxidative phosphorylation (OxPhos) and resistance to oxidative stress. Interestingly, the treatment of BRAF-mutant melanomas with BRAF inhibitors leads to addiction of tumor cells to OxPhos.Citation8
In our study, we built upon these findings for MitoBiogenesis and mechanistically dissected intrinsic therapy resistance to develop a rational approach for overcoming both intrinsic and acquired drug resistance.Citation1 We analyzed expression of the MitoBiogenesis transcriptional signature in a panel of BRAF-mutant melanomas using both cell lines and patients' tumors. We found that MAPKi downregulate MitoBiogenesis in a subset of MAPKi-sensitive cells with high basal levels of MitoBiogenesis and upregulate it in MAPKi-resistant cells with low basal levels (). These intrinsically resistant cells depend on upregulated MitoBiogenesis for survival and have increased mitochondrial DNA (mtDNA) copy number, mitochondrial mass, maximal oxygen consumption rate, and reactive oxygen species production. We were also able to validate upregulation of the MitoBiogenesis gene signature, mtDNA content, and representative OxPhos subunits in a subset of early on- and post-treatment tumor biopsies derived from BRAF-mutant melanoma patients treated with BRAF-targeting therapies.
Figure 1. Mitochondrial biogenesis (MitoBiogenesis) is a drug resistance mechanism and therapeutic target in melanoma. BRAF mutant melanoma cells with an intrinsic low basal level of MitoBiogenesis upregulate MitoBiogenesis, increase mitochondrial DNA (mtDNA) content, and alter tumor bioenergetics to survive mitogen activated protein kinase (MAPK) inhibition. Gamitrinib treatment or the knockdown of tumor necrosis factor receptor-associated protein 1 (TFAM) or mitochondrial transcription factor A (TRAP1) synergizes with MAPK inhibitors to kill resistant melanoma cells.
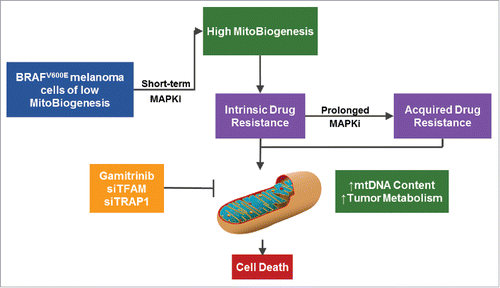
Previously, PPARGC1α was implicated as the key mediator of MitoBiogenesis in cancer cells.Citation7 We found that depletion of tumor necrosis factor receptor-associated protein 1 (TRAP1) or mitochondrial transcription factor A (TFAM), but not of PPARGC1α, substantially enhanced the efficacy of MAPK inhibition. Both TRAP1 and TFAM are important regulators of MitoBiogenesis and play roles in mitochondrial protein folding and genome replication, respectively. Our siRNA-based screening data lead us to hypothesize that gamitrinib, an inhibitor of TRAP1, would be effective in inhibiting MitoBiogenesis. Indeed, the combination of gamitrinib and MAPKi impaired MitoBiogenesis and inhibited aberrant tumor bioenergetics in vitro. The synergistic effect of the 2 inhibitors significantly suppressed tumor growth in vivo. Analyses of The Cancer Genome Atlas (TCGA) melanoma cases revealed that melanoma patients with higher expression of mitochondrial biogenesis transcriptional signature or co-expression of glycolysis and OxPhos have markedly worse overall survival rates. Thus, there is a clinical relevance to this phenotype and further investigations are warranted.
For future studies, we envision that gamitrinib will synergize with MAPKi in the treatment of BRAF-mutant melanomas with acquired drug resistance. An increase in expression of MITF and PPARGC1α in neuroblastoma RAS viral oncogene homolog (NRAS) mutant melanoma cells upon MEK inhibition has already been shown.Citation9 Thus, gamitrinib may be effective in combination with a MEK1/2 inhibitor to treat NRAS-mutant melanomas.Citation10 It is likely that the MitoBiogenesis transcriptional signature will also be modulated in tumors derived from patients who progressed on anti-CTLA4, anti-PD-1, and/or anti-PD-L1 therapies and it will be interesting to investigate whether and how MitoBiogenesis affects response and resistance to immune checkpoint blockade therapies.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Zhang G, Frederick DT, Wu L, Wei Z, Krepler C, Srinivasan S, Chae YC, Xu X, Choi H, Dimwamwa E, et al. Targeting mitochondrial biogenesis to overcome drug resistance to MAPK inhibitors. J Clin Invest. 2016 May 2; 126(5):1834–56; PMID:27043285; http://dx.doi.org/10.1172/JCI82661
- Schadendorf D, Hauschild A. Melanoma in 2013: Melanoma–the run of success continues. Nat Rev Clin Oncol 2014; 11(2):75-6; PMID:24419300; http://dx.doi.org/10.1038/nrclinonc.2013.246
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. Mutations of the BRAF gene in human cancer. Nature 2002; 417(6892):949-54; PMID:12068308; http://dx.doi.org/10.1038/nature00766
- Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 2011; 364(26):2507-16; PMID:21639808; http://dx.doi.org/10.1056/NEJMoa1103782
- Long GV, Stroyakovskiy D, Gogas H, Levchenko E, de Braud F, Larkin J, Garbe C, Jouary T, Hauschild A, Grob JJ, et al. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N Engl J Med 2014; 371(20):1877-88; PMID:25265492; http://dx.doi.org/10.1056/NEJMoa1406037
- Shi H, Hugo W, Kong X, Hong A, Koya RC, Moriceau G, Chodon T, Guo R, Johnson DB, Dahlman KB, et al. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov 2014; 4(1):80-93; PMID:24265155; http://dx.doi.org/10.1158/2159-8290.CD-13-0642
- Vazquez F, Lim JH, Chim H, Bhalla K, Girnun G, Pierce K, Clish CB, Granter SR, Widlund HR, Spiegelman BM, et al. PGC1alpha expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer Cell 2013; 23(3):287-301; PMID:23416000; http://dx.doi.org/10.1016/j.ccr.2012.11.020
- Haq R, Shoag J, Andreu-Perez P, Yokoyama S, Edelman H, Rowe GC, Frederick DT, Hurley AD, Nellore A, Kung AL, et al. Oncogenic BRAF regulates oxidative metabolism via PGC1alpha and MITF. Cancer Cell 2013; 23(3):302-15; PMID:23477830; http://dx.doi.org/10.1016/j.ccr.2013.02.003
- Gopal YN, Rizos H, Chen G, Deng W, Frederick DT, Cooper ZA, Scolyer RA, Pupo G, Komurov K, Sehgal V, et al. Inhibition of mTORC1/2 overcomes resistance to MAPK pathway inhibitors mediated by PGC1alpha and oxidative phosphorylation in melanoma. Cancer Res 2014; 74(23):7037-47; PMID:25297634; http://dx.doi.org/10.1158/0008-5472.CAN-14-1392
- Ascierto PA, Schadendorf D, Berking C, Agarwala SS, van Herpen CM, Queirolo P, Blank CU, Hauschild A, Beck JT, St-Pierre A, et al. MEK162 for patients with advanced melanoma harbouring NRAS or Val600 BRAF mutations: a non-randomised, open-label phase 2 study. Lancet Oncol 2013; 14(3):249-56; PMID:23414587; http://dx.doi.org/10.1016/S1470-2045(13)70024-X
