ABSTRACT
Improved insight into cancer cell populations responsible for treatment failure will lead to better outcomes for patients. We herein highlight a single-cell study of B-cell precursor acute lymphoblastic leukemia (BCP-ALL) at diagnosis that revealed hidden developmentally dependent cell signaling states uniquely associated with relapse.
B-cell precursor acute lymphoblastic leukemia (BCP-ALL) is the most common malignancy in childhood, arising from aberrant expansion of B cell precursors in the bone marrow. Five-year survival rates exceed 85% with modern therapies, however survival following relapse remains a significant problem and leading cause of cancer-related mortality in childhood.Citation1 Predicting patients at risk for relapse involves a combination of clinical characteristics, prognostic somatic mutations and early response to therapy.Citation2 Current treatment protocols are risk-stratified such that patients are assigned a treatment intensity based on their perceived risk of relapse. These risk prediction metrics are imperfect and more than half of relapses occur in patients not classified as high-risk.Citation3 Whereas about 2-5% of patients die due to toxicities of treatment.Citation4
There is debate as to whether resistant populations of cancer cells are present at the time of diagnosis or develop under the pressure of therapy; many studies have suggested that it is the former.Citation5,Citation6 During remission, cancer cells are rendered to an undetectable amount, consequently, if prognostically predictive populations exist, it is reasonable to assume that these cells are a rare subset of the bulk tumor cells. For this reason, high-throughput analysis of single cells in BCP-ALL patients are required to achieve a better understanding to cell types resistant to conventional therapy.
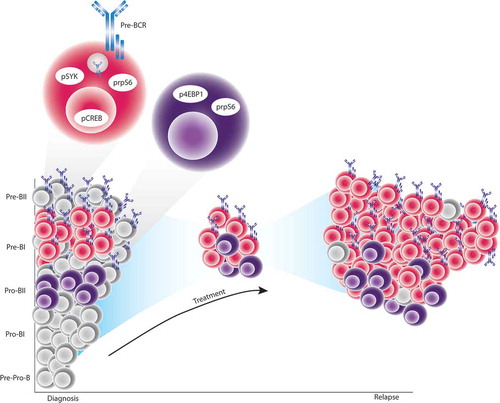
In this work, Good et al. performed single-cell studies of diagnostic leukemic samples with the goal of identifying cell populations associated with future relapse.Citation7 The authors used mass cytometry to profile 60 BCP-ALL diagnostic patient samples and 5 healthy donor samples using a 35-antibody panel which included phenotypic, functional and transcription factors involved in the development of B-cells. To overcome intratumoral and intrapatient heterogeneity, Good et al. presented an innovative approach to study BCP-ALL based on the alignment of each cancer cell to it’s most similar healthy stage of B-cell development. Using this developmental classification, the authors identified the transitional populations between the pre-pro-B and pre-BI states to be expanded in BCP-ALL patients compared to healthy donors.Citation7
Applying machine learning to the proteomic features of the expanded populations, they constructed a predictive model of relapse termed Developmentally Dependent Predictor of Relapse (DDPR) able to accurately predict time to relapse in a retrospective analysis of the cohort.Citation7 The model implicated six cellular features, confined to the pro-BII and pre-BI populations, to be associated with future relapse. In particular, patients who would go on to relapse had aberrant pre-B cell Receptor signaling assessed by increased levels of phosphorylated spleen tyrosine kinase (pSYK) and phosphorylated c-AMP responsive element-binding protein (pCREB) as well as aberrant mTOR signaling with increase expression of phosphorylated 4E binding-protein 1 (p4EBP1) and phosphorylated ribosomal protein S6 (prpS6) within these developmental subpopulations. Interestingly analysis of paired samples from 7 patients at diagnosis and relapse revealed the persistence of these cells and their predictive features at the relapse. Excitingly, using DDPR as a clinical risk prediction tool, it performed well to identify patients at high risk for future relapse but moreover, DDPR augmented the prediction power of currently used methods of clinical risk prediction. This suggests utility in patient care if DDPR is able to be more widely adopted.
Together these data suggest that the pre-pro-B to pre-BI transition is the most vulnerable to malignant transformation in BCP ALL. It has been previously demonstrated that across these developmental populations in healthy B-cells, two important coordination points occur: the pro-B and pre-B checkpoints.Citation8 At the pro-B checkpoint, signaling through the IL-7 receptor via the JAK/STAT and PI3K/mTOR pathways enforces pro-survival signals as B-cells attempt to rearrange the immunoglobulin heavy chain. The second coordination point occurs following heavy chain rearrangement to determine if it was productive and if they may proceed through differentiation. This checkpoint involves signaling through the pre-B cell receptor.Citation8
DDPR predictive features suggested that patients who will relapse have aberrant activation of the signaling networks important in cells phenotypically similar to those at the coordination points.Citation7 These cells are resistant to conventional chemotherapy and are enriched, specifically for the pre-BI cells, at time of relapse (). The signaling phenotypes identified by DDPR are potentially targetable using PI3K/mTOR inhibition and ABL/SRC inhibition. More mechanistic studies will inform how BCP-ALL exploits these pathways to understand why these developmental cell types are vulnerable to leukemogenesis and treatment resistance. Understanding such mechanisms will allow integration of the biologic and clinical parameters to identify optimal treatment for each patient. Finally, the performance of DDPR is promising to improve risk stratification and treatment decisions for patients.
Figure 1. Single-cell developmental classification and DDPR prediction to model relapse in BCP-ALL.
At diagnosis, expanded leukemic cells have the closest phenotypic similarity to cells across the pre-pro-B to pre-BI transitional populations of normal B-cell development. BCP-ALL cells with distinct cellular features and the highest phenotypic similarity to pro-BII and pre-BI cells exist at diagnosis in patients who will go on to relapse. Specifically, pro-BII-like cells with high basal activation of either prpS6 or 4EBP1, and pre-BI-like cells with high basal activation of SYK and lack of CREB and rpS6 response to pre-B cell receptor engagement. These cells exist at diagnosis but persist despite the pressure of treatment to mediate eventual relapse.
Additional information
Funding
References
- Mullighan CG. Molecular genetics of B-precursor acute lymphoblastic leukemia. J Clin Invest. 2012;122(10):3407–3415. doi:10.1172/JCI61203.
- Conter V, Bartram CR, Valsecchi MG, Eriksson N, Crawford DC, Lee M-TM, Chen C-H, Motsinger-Reif A, Sagreiya H, Liu N, et al. Molecular response to treatment redefines all prognostic factors in children and adolescents with B-cell precursor acute lymphoblastic leukemia: results in 3184 patients of the AIEOP-BFM ALL 2000 study. Blood. 2010;115(16):3206–3214. doi:10.1182/blood2009-10-248146.
- Prucker C, Attarbaschi A, Peters C, Dworzak MN, Po U. Induction death and treatment-related mortality in first remission of children with acute lymphoblastic leukemia: a population-based analysis of the Austrian Berlin-Frankfurt-Münster study group. Leukemia. 2009 Jul;23(7):1264–9. doi:10.1038/leu.2009.12.
- Anderson K, Lutz C, Van Delft FW, Bateman CM, Guo Y, Colman SM, Kempski H, Moorman AV, Titley I, Swansbury J, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469(7330):356–361. doi:10.1038/nature09650.
- Longo DL, Hunger SP, Mullighan CG. Acute lymphoblastic leukemia in children. N Engl J Med. 2015;373(16):1541–1552. doi:10.1056/NEJMra1400972.
- Good Z, Sarno J, Jager A, Samusik N, Aghaeepour N, Simonds EF, White L, Lacayo NJ, Fantl WJ, Fazio G, et al. Single-cell developmental classification of B cell precursor acute lymphoblastic leukemia at diagnosis reveals predictors of relapse. Nat Med. 2018;24(January):474–483. doi:10.1038/nm.4505.
- Bendall SC, Davis KL, Amir EAD, et al. Single-cell trajectory detection uncovers progression and regulatory coordination in human b cell development. Cell. 2014;157(3):714–725. doi:10.1016/j.cell.2014.04.005.
- Müschen M. Autoimmunity checkpoints as therapeutic targets in B cell malignancies. Nat Rev Cancer. 2018;18(2):103–116. doi:10.1038/nrc.2017.111.
