ABSTRACT
Chromatin remodeling is critical for the regulation of the DNA damage response. We highlight findings from our recent study showing that the deposition of the histone variant H3.3 by the alpha-thalassemia mental retardation X-linked protein (ATRX) and the death domain associated protein (DAXX) chromatin remodeling complex regulates DNA repair synthesis during homologous recombination.
Chromatin modification and remodeling is a dynamic process that is established not only as prerequisite to allow access to DNA for repair following damage, but also as a key regulator of the DNA damage response (DDR). Chromatin dynamics during the DDR involve histone modifications, such as phosphorylation and methylation, and also the disassembly and reassembly of nucleosomes before and after DNA repair, respectively. The removal of original histones is often followed by the deposition of histone variants that play important roles in the stress response. Extensive efforts have been dedicated to identify the functions of histone variants in the DDR and the different repair mechanisms. Among the most critical lesions occurring in the cell are DNA double strand breaks (DSBs), which can be repaired by 2 major pathways: non-homologous end-joining (NHEJ) and homologous recombination (HR). HR can be regulated by the early deposition and removal of the histone variant H2A.Z, which has been shown to control resection, an important processing step on break ends that commits the cell to repair by HR.Citation1 Another variant, H3.3, has been shown to be incorporated rapidly after DNA damage and promotes NHEJ.Citation2 It is also required for post-repair chromatin assembly at sites of DSBs repaired by NHEJ.Citation3 While it is clear that histone exchange can regulate early DSB repair steps, such as repair pathway choice, chromatin dynamics influencing later stages, particularly in HR, remain poorly characterized.
One distinction between NHEJ and HR is the requirement for a sister chromatid in HR which is used as a template for repair synthesis to allow the faithful restoration of the original sequence. Repair synthesis occurs in various repair mechanisms, such as nucleotide excision repair (NER), but its regulation during HR is not well understood in human cells.Citation4 While it has been shown that repair synthesis in NER is concomitant with chromatin reconstitution through the chromatin assembly factor-1 (CAF-1), whether a similar process occurs in HR has not been shown.Citation5 In our recently published work, we describe a novel role for H3.3 deposition by the chromatin remodeler alpha-thalassemia mental retardation X-linked protein (ATRX) and the chaperon death domain associated protein (DAXX) in a sub-pathway of HR-mediated DSB repair.Citation6 We show that cells lacking ATRX, DAXX or H3.3 have a defect in repairing ionizing radiation-induced DSBs by HR. Strikingly, cells lacking these factors exhibit a strong defect in extended DNA repair synthesis, indicating a role of the chromatin remodeling complex during this late HR step. We employed a system for the in vivo visualization of H3.3 dynamics and showed that newly synthesized H3.3 is incorporated at sites of DNA damage at both early and late time points. However, only late incorporation was ATRX dependent and was associated with DNA synthesis. To further consolidate the association of repair synthesis with nucleosome assembly, we identified a strong damage-induced interaction between ATRX and proliferating cell nuclear antigen, which is required for both repair synthesis and repair completion.
Our findings inspire a model in which the chromatin remodeler ATRX promotes extended DNA synthesis by simultaneously facilitating chromatin reconstitution. Unlike in NER, this is a prerequisite for repair completion as the disruption of this process impedes repair. One possible explanation for this discrepancy is the extent of repair synthesis, which is much longer during HR and possibly requires nucleosome reassembly to allow further progression of the DNA synthesis machinery. A tight coupling of histone deposition with DNA synthesis also occurs during replication, where the coordination of chromatin assembly and DNA synthesis is important for the regulation of elongation rate and fork progression.Citation7 Additionally, histone deposition is important during transcription and can influence transcription rate.Citation8 Therefore, histone exchange seems to be critical for the regulation of processes that require extended DNA/RNA synthesis such as replication, transcription and repair (). This is not surprising given the importance of maintaining genomic integrity in processes exposing naked DNA that needs protection, especially over long patches, a notion in agreement with the described gap-filling role of H3.3 during transcription.Citation9 Furthermore, chromatin reconstitution can also serve a structural stabilization function to alleviate topological stress occurring during DNA unwinding, a major impediment for various DNA/RNA synthesis processes.
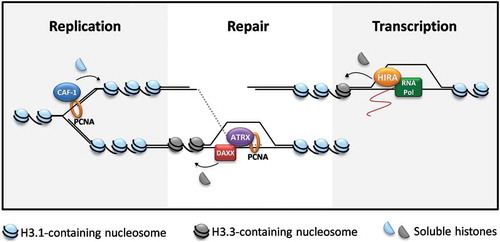
Figure 1. Histone dynamics in replication, repair synthesis and transcription. Chromatin remodeling is a common requirement for the regulation of DNA and RNA synthesis. Distinct chromatin remodelers and chaperons direct the deposition of histone variants during replication (chromatin assembly factor-1, CAF-1), repair synthesis in homologous recombination (alpha-thalassemia mental retardation X-linked protein, ATRX, and the chaperon death domain associated protein, DAXX) and transcription (histone regulator A, HIRA). PCNA: proliferating nuclear cell antigen; RNA Pol: RNA polymerase II.
The role of ATRX during HR provides insight into how DNA repair synthesis is regulated in vivo and highlights the importance of histone variants and their remodelers in this process. It would be of interest to study similar processes in other repair pathways, which would establish a paradigm for chromatin reorganization during DNA repair synthesis. Additionally, this newly identified role for ATRX could help to better understand the diseases in which ATRX, H3.3 or DAXX are mutated. One prominent example is a subset of cancers that undergo a telomerase-independent telomere maintenance process known as the alternative lengthening of telomeres (ALT). ALT cancers represent about 10–15% of all cancers and almost always have ATRX, H3.3 or DAXX mutations.Citation10 ALT is identified as an HR process that involves long range break-induced DNA synthesis. Given that ATRX’s function is lost during ALT, it was surprising to uncover its HR role which raises the question of how this new role fits to the ALT process. One possibility is that ATRX has distinct functions at telomeres versus internal breaks. Alternatively, ATRX might promote a form of HR that is repressive to ALT and its loss allows HR that is more permissive to the ALT process. Delineating these functions is important for the understanding of ALT and can be of value for the development of therapeutic strategies for ALT cancers.
Disclosure of interest
The authors report no conflict of interest.
Additional information
Funding
References
- Gursoy-Yuzugullu O, Ayrapetov MK, Price BD. Histone chaperone Anp32e removes H2A.Z from DNA double-strand breaks and promotes nucleosome reorganization and DNA repair. Proc Natl Acad Sci USA. 2015;112:7507–7512. PMID:26034280. doi:10.1073/pnas.1504868112.
- Luijsterburg MS, De Krijger I, Wiegant WW, Shah RG, Smeenk G, De Groot AJL, Pines A, Vertegaal ACO, Jacobs JJL, Shah GM, et al. PARP1 links CHD2-mediated chromatin expansion and H3.3 deposition to DNA repair by non-homologous end-joining. Mol Cell. 2016;61:547–562. PMID:26895424. doi:10.1016/j.molcel.2016.01.019.
- Li X, Tyler JK. Nucleosome disassembly during human non-homologous end joining followed by concerted HIRA- and CAF-1-dependent reassembly. Elife. 2016;5:e15129. PMID: 7269284. doi:10.7554/eLife.15129.001.
- Wright WD, Shah SS, Heyer WD. Homologous recombination and the repair of DNA double-strand breaks. J Biol Chem. 2018 Jul;27:10524–10535. PMID:29599286. doi:10.1074/jbc.TM118.000372.
- Polo SE, Roche D, Almouzni G. New histone incorporation marks sites of UV repair in human cells. Cell. 2006;127:481–493. PMID:17081972. doi:10.1016/j.cell.2006.08.049.
- Juhász S, Elbakry A, Mathes A, Löbrich M. ATRX promotes DNA repair synthesis and sister chromatid exchange during homologous recombination. Mol Cell. 2018;71:11–24. PMID:29937341. doi:10.1016/j.molcel.2018.05.014.
- Kurat CF, Yeeles JT, Patel H, Early A, Diffley JF. Chromatin controls DNA replication origin selection, lagging-strand synthesis, and replication fork rates. Mol Cell. 2017;65:117–130. PMID:27989438. doi:10.1016/j.molcel.2016.11.016.
- Sarai N, Nimura K, Tamura T, Kanno T, Patel MC, Heightman TD, Ura K, Ozato K. WHSC1 links transcription elongation to HIRA-mediated histone H3.3 deposition. EMBO J. 2015;32:2392–2406. PMID:23921552. doi:10.1038/emboj.2013.176.
- Ray-Gallet D, Woolfe A, Vassias I, Pellentz C, Lacoste N, Puri A, Schultz DC, Pchelintsev NA, Adams PD, Jansen LE, et al. Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol Cell. 2011;44:928–941. PMID:22195966. doi:10.1016/j.molcel.2011.12.006.
- Dilley RL, Greenberg RA. ALTernative telomere maintenance and cancer. Trends Cancer. 2015;1:145–156. PMID:26645051. doi:10.1016/j.trecan.2015.07.007.
