ABSTRACT
The search for mechanisms underlying different cellular responses to the treatment with Nutlin-3, an MDM2 inhibitor that unleashes p53, revealed a translational control mechanism involving the RNA binding proteins PCBP2 and, particularly, DHX30. Sifting through a multi-functional p53-dependent transcriptional output, this translational control can modulate the activation of cell death pathways.
The long list of titles the TP53 protein (best known and hereinafter referred to as p53) collected during the years testifies its complex, in some ways unprecedented, relevance in many cellular pathways.Citation1 But p53, the guardian of the genome, puzzle or paradigm, good cop, jack of all trades, guardian of immunity, master regulator, is still hiding critical aspects of its functions.Citation2,Citation3
Although non-cell autonomous, and non-nuclear roles have been described, p53 mainly functions as a sequence-specific transcription factor. The p53 binding sites in promoters and enhancer regions have been annotated, cataloged, and dissected in hundreds of studies defining a core group of direct p53 target genes.Citation4 This group comprises at least 150 genes involved in different functions, ranging from cell cycle arrest to DNA repair, cell death, differentiation, and senescence. Hence, a persistent question in the field is to identify the bases for transactivation specificity/selectivity that would match and predict a specific cell outcome in response to p53-induced transcriptional changes. This question is still open, despite the discovery of many features, such as the quality of p53 binding sites, the effect of post-translational modifications, the role of specific cofactors, and the crosstalk with other transcription factors.Citation3 The multiplicity and complexity of the various upstream controls and downstream feedback loops tuning p53 functions is nothing short of astounding. The most prominent of such negative feedback loops to have been discovered involves MDM2 (Mouse Double Minute 2), an E3 ubiquitin ligase whose expression is stimulated by p53 and that promotes p53 degradation.Citation5
Initially, the discovery of many microRNAs and long noncoding RNAs whose transcription is directly modulated by p53, has brought light to the fact that p53-directed functions can merge transcriptional and post-transcriptional controls. Indeed, we now appreciate that it is the combination of these regulatory layers that eventually shapes the cellular fate. Our understanding of how p53 can act in post-transcriptional and translational control has been recently increased by the identification of direct and indirect modulation of RNA-binding proteins, translation initiation factors, and elements of ribosome biogenesis.Citation6
In a first attempt to focus on translation to interpret p53-mediated responses, we used polysomal profiling to study translatome changes, i.e. the modulation of relative mRNA association with actively translating ribosomes. We compared it to total RNA, i.e. the typical transcriptome view, to examine the consequences of p53 activation via DNA damage response (doxorubicin treatment) or MDM2 inhibition (Nutlin-3 treatment).Citation7 We surprisingly observed a high level of uncoupling, i.e. changes occurring in the translatome without a corresponding change in the transcriptome, as well as changes in the relative translation efficiency for specific transcripts.
However, we realized the strong impact of the translation uncoupling on cell fate only after exploring the translation of cell lines undergoing opposite cellular outcomes upon p53 activation, namely cell cycle arrest or apoptosis. Indeed, transcriptional control is not diverse enough to predict the markedly opposite cellular responses to the p53 activation.Citation8
Translatome analysis revealed that multiple cell lines undergoing apoptosis consistently upregulate the translation of a unique class of transcripts enriched for apoptosis function. This class was exclusively associated with a highly enriched sequence motif located in 3ʹ untranslated regions (3ʹUTRs). We labeled this motif CGPD (for CG-rich-motif for p53 dependent death). We confirmed that it was sufficient for translational upregulation of a reporter gene in pro-apoptotic cells, but not in cells undergoing cell cycle arrest, and that its effect required wild-type p53 expression.Citation9
We identified several RNA-binding proteins (RBPs) with the potential to interact with the CGPD motif. Among these, Poly(RC) Binding Protein 2 (PCBP2) and DExH-Box Helicase 30 (DHX30) were found only in cell cycle arrested cells. Hence, we demonstrated that the cooperation of these two RBPs limits the translation potential of pro-apoptotic targets in the context of p53-dependent apoptosis.Citation9 As a consequence, the transient or constitutive depletion of DHX30 can lead cell cycle arrested cells to partially phenocopy apoptotic cells. The converse was instead observed when DHX30 was over-expressed in apototic-prone cells.
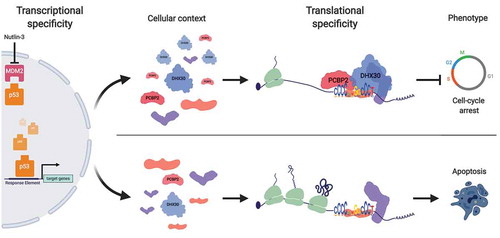
Thus, our work identifies translational specificity as a critical regulatory layer for cancer cell proneness to undergo cell death downstream of a p53-activating treatment (). One such regulatory mechanism involves PCBP2 and, particularly, DHX30 acting through a target cis-element in 3ʹUTRs.
Figure 1. The RBP repertoire modulates p53-dependent cell fate. The p53 sequence-specific transcription factor controls a vast network of direct targets genes by binding to variations of Response Element (RE) binding sites. This multifunctional p53-transcriptional network can be modulated at the translational level, for example by PCBP2 and DHX30 targeting the CGPD-motif depending on their relative levels of expression, to influence cell fate.
Several open questions remain. Which is the exact mechanism by which DHX30 can control sequence-specific translation? Can we exploit DHX30 levels to predict the outcome of treatments activating p53? Finally, it will be essential to understand the specific role p53 plays in this translation control mechanism. It may even be that, adapting the title of an insightful review,Citation10 p53 is a jack of all trades, and a master of one.
References
- Lane D, Levine A. p53 research: the past thirty years and the next thirty years. Cold Spring Harb Perspect Biol. 2010;2:1. doi:10.1101/cshperspect.a000893.
- Ko LJ, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–2. doi:10.1101/gad.10.9.1054.
- Kastenhuber ER, Lowe SW. Putting p53 in context. Cell. 2017;170:1062–1078. doi:10.1016/j.cell.2017.08.028.
- Nguyen T-AT, Grimm SA, Bushel PR, Li J, Li Y, Bennett BD, Lavender CA, Ward JM, Fargo DC, Anderson CW, et al. Revealing a human p53 universe. Nucleic Acids Res. 2018;46:8153–8167. doi:10.1093/nar/gky720.
- Kubbutat MHG, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi:10.1038/387299a0.
- Marcel V, Catez F, Diaz JJ. p53, a translational regulator: contribution to its tumour-suppressor activity. Oncogene. 2015;34:5513–5523. doi:10.1038/onc.2015.25.
- Zaccara S, Tebaldi T, Pederiva C, Ciribilli Y, Bisio A, Inga A. p53-directed translational control can shape and expand the universe of p53 target genes. Cell Death Differ. 2014;21:1522–1534. doi:10.1038/cdd.2014.79.
- Andrysik Z, Galbraith MD, Guarnieri AL, Zaccara S, Sullivan KD, Pandey A, MacBeth M, Inga A, Espinosa JM. Identification of a core TP53 transcriptional program with highly distributed tumor suppressive activity. Genome Res. 2017;27:1645–1657. doi:10.1101/gr.220533.117.
- Rizzotto D, Zaccara S, Rossi A, Galbraith MD, Andrysik Z, Pandey A, Sullivan KD, Quattrone A, Espinosa JM, Dassi E, et al. Nutlin-induced apoptosis is specified by a translation program regulated by PCBP2 and DHX30. Cell Rep. 2020;30:1–15. doi:10.1016/j.celrep.2020.03.011.
- Junttila MR, Evan GI. P53 a Jack of all trades but master of none. Nat Rev Cancer. 2009;9:821–829. doi:10.1038/nrc2728.
