ABSTRACT
Circular RNAs (circRNAs) are expressed and are regulated in many biological processes but little is known about their ability to directly control mRNA homeostasis. We show that circRNA zinc finger protein 609 (circZNF609) interacts with several mRNAs increasing the final protein levels, which in the case of the cytoskeleton-associated protein 5 (CKAP5) leads to a stabilized microtubule cytoskeleton and an enhanced tumor cell proliferation.
The latest research showed the importance of RNA molecules as a key factor in the regulatory circuitries. To carry out these fine-tuning activities, RNA not only interacts with proteins and chromatin but often directly pairs with other RNA molecules.Citation1,Citation2 In addition to the well-known activity of microRNAs (miRNAs) on mRNAs, much interest is now being directed to the study of how long non-coding RNAs (lncRNAs) control gene expression by targeting specific mRNAs to regulate their stability, translation, and localization by pairwise interaction. Within this frame of research, growing interest is dedicated to circular RNAs (circRNAs), a well-established class of RNA molecules originating from a back-splicing event in which a downstream splice-donor site is joined to an upstream splice-acceptor site, yielding a covalently closed circular RNA. CircRNAs show several peculiar features, such as evolutionary conservation and tissue-specific expression, but above all, they have been found to be deregulated in many pathological conditions, including cancer. Several studies indicate that circRNAs elicit their function as miRNA sponges, but the cell-type-specific microRNA signature restrains this mechanism as a general mode of action. In our workCitation3we explored the regulatory roles of the RNA-RNA interactions involving the circRNA zinc finger protein 609 (circZNF609), a circRNA previously reported to be regulated and to control human primary myoblast and embryonal rhabdomyosarcoma (ERMS) cell proliferation.Citation4,Citation5 CircZNF609 was also found to be upregulated in several types of human cancers, such as prostate cancer, breast cancer, gastric cancer, nasopharyngeal carcinoma, and hepatocellular carcinoma, where its depletion has been linked with reduced aggressiveness of tumor cells.
Using a psoralen-crosslinking RNA pulldown to detect RNA-RNA interactions in vivo, we found that circZNF609 pairs directly with a few mRNAs, among which is the mRNA of the cytoskeleton-associated protein 5 (CKAP5). A more detailed molecular study points to the back-splicing junction of circZNF609 as the sequence responsible for this RNA-RNA pairing, underlining the specificity of this interaction in front of the linear ZNF609 mRNA counterpart. As a result, the CKAP5 mRNA is stabilized and its translation is enhanced. We demonstrated that this stabilization is produced by ELAV like RNA Binding Protein 1 (ELAVL1, also known as HuR), an RNA-binding protein (RBP) known to induce mRNA stabilization and translation and to control lncRNA metabolism.Citation6,Citation7 Specifically, circZNF609 contains several-binding sites for ELAVL1 in its sequence, and the interaction between the circRNA and CKAP5 mRNA will facilitate ELAVL1 loading from the circular onto the messenger RNA ().
Figure 1. The potential mechanism through which circRNA zinc finger protein 609(circZNF609) regulates mRNA stability. CircZNF609 contains several binding sites for ELAV like RNA binding protein 1 (ELAVL1). The circRNA interaction with the cytoskeleton-associated protein 5 (CKAP5) transcript favours the binding of ELAVL1 to the mRNA, increasing mRNA levels and translation. Lack of circZNF609 or disruption of the circZNF609-mRNA interaction will impair mRNAstability and translation of CKAP5 protein leading to altered microtubules (MT)dynamics.
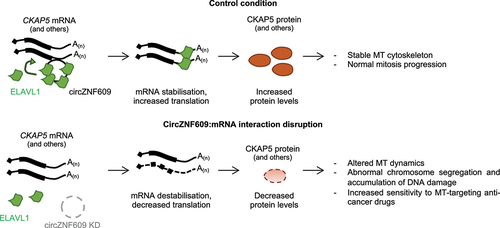
The CKAP5 protein is a highly conserved cytoskeleton-associated factor regulating several aspects of microtubule function: it promotes microtubule nucleation, binds to their growing (“plus”) ends, sustains their elongation and regulates their dynamics.Citation8 It also controls mitotic spindle formation and chromosome segregation by stabilizing kinetochore fibers, thus having an important role in mitotic progression.Citation9 We demonstrated that the interference of circZNF609, and hence the downregulation of CKAP5 protein levels, produces a de-regulation of microtubule dynamics, leading to abnormal chromosome segregation and DNA damage accumulation, which will eventually produce the cell-cycle halt previously observed in rhabdomyosarcoma.Citation4
Microtubule dynamics is essential to all processes depending on the cytoskeleton, including cell migration and differentiation, the building-up of the mitotic scaffold and chromosome segregation. Several first-line chemotherapeutic agents, such as vincristine or Paclitaxel/Taxol, are designed to target either the microtubule assembly from tubulin or their dynamic activity; they are effective chemotherapeutic agents and are still in use to treat cancer of various origins, including rhabdomyosarcoma. We demonstrated that preventing the circZNF609/CKAP5 mRNA interaction, through either small interfering RNAs (siRNAs) against circZNF609 or locked nucleic acid (LNA)-modified oligonucleotides against the pairing region, sensitizes rhabdomyosarcoma cells to microtubule-targeting chemotherapeutic agents. Moreover, we demonstrated that this regulatory mechanism is also conserved in other cancer cell models such as neuroblastoma and chronic myelogenous leukemia, setting such blockage as a potential therapeutic coadjuvant in cancer treatment.
Microtubule dynamics is essential to all processes depending on the cytoskeleton, including cell migration and differentiation, the building-up of the mitotic scaffold and chromosome segregation. Several first-line chemotherapeutic agents, such as vincristine or Paclitaxel/Taxol, are designed to target either the microtubule assembly from tubulin or their dynamic activity; they are effective chemotherapeutic agents and are still in use to treat cancer of various origins, including rhabdomyosarcoma. We demonstrated that preventing the circZNF609/CKAP5 mRNA interaction, through either small interfering RNAs (siRNAs) against circZNF609 or locked nucleic acid (LNA)-modified oligonucleotides against the pairing region, sensitizes rhabdomyosarcoma cells to microtubule-targeting chemotherapeutic agents. Moreover, we demonstrated that this regulatory mechanism is also conserved in other cancer cell models such as neuroblastoma and chronic myelogenous leukemia, setting such blockage as a potential therapeutic coadjuvant in cancer treatment.
Microtubule dynamics is essential to all processes depending on the cytoskeleton, including cell migration and differentiation, the building-up of the mitotic scaffold and chromosome segregation. Several first-line chemotherapeutic agents, such as vincristine or Paclitaxel/Taxol, are designed to target either the microtubule assembly from tubulin or their dynamic activity; they are effective chemotherapeutic agents and are still in use to treat cancer of various origins, including rhabdomyosarcoma. We demonstrated that preventing the circZNF609/CKAP5 mRNA interaction, through either small interfering RNAs (siRNAs) against circZNF609 or locked nucleic acid (LNA)-modified oligonucleotides against the pairing region, sensitizes rhabdomyosarcoma cells to microtubule-targeting chemotherapeutic agents. Moreover, we demonstrated that this regulatory mechanism is also conserved in other cancer cell models such as neuroblastoma and chronic myelogenous leukemia, setting such blockage as a potential therapeutic coadjuvant in cancer treatment.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7–3. doi:10.1038/nrg3606.
- Statello L, Guo C-J, Chen L-L, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22(2):96–118. doi:10.1038/s41580-020-00315-9.
- Rossi F, Beltran M, Damizia M, Grelloni C, Colantoni A, Setti A, Di Timoteo G, Dattilo D, Centrón-Broco A, Nicoletti C. Circular RNA ZNF609/CKAP5 mRNA interaction regulates microtubule dynamics and tumorigenicity. Mol Cell. 2022;82(1):75–89.e9. doi:10.1016/j.molcel.2021.11.032.
- Rossi F, Legnini I, Megiorni F, Colantoni A, Santini T, Morlando M, Di Timoteo G, Dattilo D, Dominici C, Bozzoni I. Circ-ZNF609 regulates G1-S progression in rhabdomyosarcoma. Oncogene. 2019;38(20):3843–54. doi:10.1038/s41388-019-0699-4.
- Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66(1):22–37.e9. doi:10.1016/j.molcel.2017.02.017.
- Galbaán S, Kuwano Y, Pullmann R, Martindale JL, Kim HH, Lal A, Abdelmohsen K, Yang X, Dang Y, Liu JO. RNA-binding proteins HuR and PTB promote the translation of hypoxia-inducible factor 1α. Mol Cell Biol. 2008;28(1):93–107. doi:10.1128/mcb.00973-07.
- Mazan-Mamczarz K, et al. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proceedings of the National Academy of Sciences of the United States of America; 2003 doi:10.1073/pnas.1432104100.
- Brouhard GJ, Stear JH, Noetzel TL, Al-Bassam J, Kinoshita K, Harrison SC, Howard J, Hyman AA. XMAP215 is a processive microtubule polymerase. Cell. 2008;132(1):79–88. doi:10.1016/j.cell.2007.11.043.
- Miller MP, Asbury CL, Biggins S. A TOG protein confers tension sensitivity to kinetochore-microtubule attachments. Cell. 2016;165(6):1428–39. doi:10.1016/j.cell.2016.04.030.
