Abstract
Background: Early detection of bacteria and their antibiotic susceptibility patterns are critical to guide therapeutic decision-making for optimal care of septic patients. The current gold standard, blood culturing followed by subculture on agar plates for subsequent identification, is too slow leading to excessive use of broad-spectrum antibiotic with harmful consequences for the patient and, in the long run, the public health. The aim of the present study was to assess the performance of two commercial assays, QuickFISH® (OpGen) and Maldi Sepsityper™ (Bruker Daltonics) for early and accurate identification of microorganisms directly from positive blood cultures.
Materials and methods: During two substudies of positive blood cultures, the two commercial assays were assessed against the routine method used at the clinical microbiology laboratory, Unilabs AB, at Skaraborg Hospital, Sweden.
Results: The Maldi Sepsityper™ assay enabled earlier microorganism identification. Using the cut-off for definite species identification according to the reference method (>2.0), sufficiently accurate species identification was achieved, but only among Gram-negative bacteria. The QuickFISH® assay was time-saving and showed high concordance with the reference method, 94.8% (95% CI 88.4–98.3), when the causative agent was covered by the QuickFISH® assay.
Conclusions: The use of the commercial assays may shorten the time to identification of causative agents in bloodstream infections and can be a good complement to the current clinical routine diagnostics. Nevertheless, the performance of the commercial assays is considerably affected by the characteristics of the causative agents.
Introduction
Sepsis is the primary cause of death from severe infections. Globally, an estimated 18 million people die from sepsis annually [Citation1]. Early sepsis diagnosis and targeted antimicrobial therapy can reduce the length of hospital stay of the patients and thereby decrease health care costs by approximately 30% [Citation2]. The current gold standard for sepsis diagnosis, blood culturing, takes 12–72 h to detect microorganisms in the blood, and even longer to identify the specific organism and its antimicrobial susceptibility to be used for optimal therapy [Citation3,Citation4]. Thus, there are demands for molecular assays that can easily be taken into the clinical microbiology laboratory routine handling, empowering earlier identification of the causative agent and its antibiogram. As the supplemented components in blood culture media, needed for antimicrobial neutralization are substituted with adsorbent polymeric beads, various molecular assays could improve by sampling directly from the positive blood culture, such as the matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) and the peptide nucleic acid (PNA) fluorescent in situ hybridization (FISH) technique.
The PNA FISH method has been applied in clinical microbiology laboratories for over 15 years for identification of a variety of organisms using fluorescent PNA probes targeting the 16S rRNA of the bacteria and 18S rRNA for fungi [Citation5–8]. This technique requires prior Gram staining and subsequently the slide is viewed under a fluorescence microscope, turn-around time 1.5–3 h. Recent introduction of the next-generation test, the QuickFISH®, the time for processing has been further reduced and the turn-around time is currently around 0.5 h. A number of different QuickFISH® assays have been developed and validated, each capable of detecting up to a maximum of three different species per assay. Staphylococcus QuickFISH® differentiates Staphylococcus aureus from coagulase-negative staphylococci (CoNS) [Citation9–12], Enterococcus QuickFISH® for identification of Gram-positive cocci in cluster differentiates Enterococcus faecalis from other Enterococcus spp., including Enterococcus faecium [Citation12–14], the Gram-Negative QuickFISH® which differentiates Escherichia coli from Pseudomonas aeruginosa and Klebsiella pneumonia [Citation12], and finally the Candida QuickFISH® assay differentiating Candida albicans from Candida glabrata and Candida parapsilosis [Citation15].
In contrast to QuickFISH®, the MALDI-TOF MS theoretically has the potential to identify any cultured microorganism from a positive blood culture [Citation16,Citation17]. Several methods for direct bacterial identification in positive blood cultures within 1 h with MALDI-TOF MS have been developed and recently summarized in a review by Dubourg et al [Citation18]. Nevertheless, this application demands removal of human cells from the specimen to eliminate interference with human proteins in the final MALDI-TOF MS analysis. The commercial Maldi Sepsityper™ kit allows that, by involving the lysis of human blood cells, followed by centrifugation and washing steps. The final result is a pellet of bacteria or fungi, which is further processed by standard methods for species identification using MALDI-TOF MS [Citation19].
In the current study, we aim to extend our earlier evaluation [Citation4] with two additional commercial assays, the Maldi Sepsityper™ and the QuickFISH® assays for early and accurate identification of microorganisms directly from positive blood cultures. The commercial assays were compared to routine diagnostics at Unilabs, the clinical microbiology laboratory at Skaraborg Hospital, Sweden, by assessing concordance of identified microorganisms between the commercial assays and the reference method, identification rates as well as estimated turnaround times.
Material and methods
Settings
The Department of Clinical Microbiology, Unilabs AB, at Skaraborg Hospital, Skövde, Sweden, receives samples from all hospital departments, which in 2015 accounted for ∼12,000 blood culture bottles from ∼5000 patients.
Study design
The commercial assays were assessed against the reference method, described below. The present study was conducted as two substudies of positive blood cultures, February–March 2013 and April–May 2015. During the first substudy, a total of 179 positive blood culture bottles were identified in the clinical laboratory. To compare the identification rate, all positive blood cultures were analysed with the Maldi Sepsityper™ assay and routine analysis using the reference method (). For 100 of the 179 positive blood cultures, the reference method reported final species identification to the clinicians. Species identification of the Maldi Sepsityper™ assays directly on these 100 positive blood cultures were compared to the reported final species identification, to assess the performance of the Maldi Sepsityper™ assay. By Gram staining of these 179 blood cultures, Gram-positive cocci in cluster were identified and selected for species identification with either the Staphylococcus and/or Enterococcus QuickFISH® assays. The assessment of the Gram-negative QuickFISH® assay was performed during the second substudy, when 203 positive blood culture bottles were identified in the clinical laboratory. By Gram staining of these blood cultures, Gram-negative bacilli were identified and selected for species identification with the Gram-negative QuickFISH® (). To assess the performance of the QuickFISH® assays, species identification by the QuickFISH® was compared to species identification reported as final result by the reference method. The species identification by the reference method was performed without knowledge of the results using the two commercial assays. Turnaround times were estimated by the laboratory personnel performing the commercial assays and the reference method.
Figure 1. Workflows and turnarounds time for the methods used in the present study. During the first substudy (a), both commercial assays, the Maldi SepsityperTM kit (i) with or (ii) without extra formic acid treatment, and the QuickFISH® test were compared with the reference method. Gram-positive cocci in clusters were selected for evaluation of the Staphylococcus and/or Enterococcus QuickFISH® tests. During the second substudy (b), only the QuickFISH® test was compared with the reference method. Gram-negative bacilli were selected for evaluation of Gram-Negative QuickFISH® test.
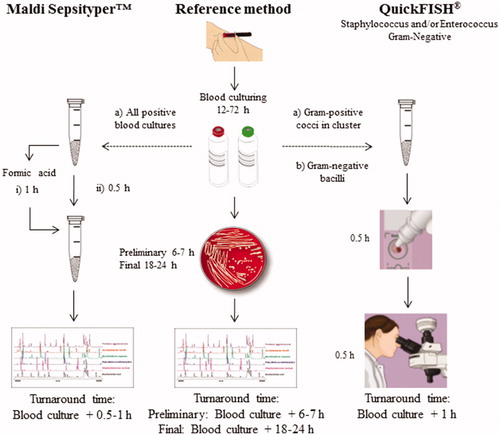
Reference method – routine diagnostics
One or two sets of blood cultures from two different puncture sites were collected for each patient according to routine clinical practice. Eight to ten millilitres whole blood was inoculated in each BacT/ALERT® FAN Plus bottle (bioMériuex, Marcy-l‘Etoile, France). Each set consisted of one aerobic and one anaerobic bottle. Blood culture processing was performed using the BacT/ALERT® FN (bioMérieux, Marcy-l‘Etoile, France). All bottles indicating microbial growth were removed from the instrument and an aliquot was taken for Gram stain by conventional methods. Results from Gram staining guided which positive blood cultures that were to be further cultured on solid media for subsequent analysis. When two or more positive blood culture bottles from the same patient showed similar Gram staining results, only one of these positive blood cultures bottles was further cultured on solid media. Definite species identification with MALDI-TOF MS was performed on a Microflex LT mass spectrometer (Bruker Daltonics, Leipzig, Germany) with BioTyper software v2.0 using default parameter settings. As recommended by Bruker Daltonics, spectral scores 2.0–2.29 was considered to be a high probability that the genus had been identified and the species was accurate. Spectral scores 1.7–1.99 was considered reliable for identification at the genus level. A score lower than 1.7 was considered as a negative result reported as ‘no reliable identification’. The species or genus with the highest score was considered to be the identified organism in the sample and reported as final result in clinical routine. Antibiotic susceptibility was determined by accredited laboratory methods according to EUCAST guidelines (www.eucast.org). Preliminary results, i.e. species identification by MALDI-TOF MS and antibiotic susceptibility on isolates after a short incubation time on solid plates, were reported to clinicians within 6–7 h, whereas final results usually were reported to clinicians within 18–24 h. For this study, only the final results, i.e. final species identification and antibiotic susceptibility determined by the reference method, were used for comparison with the two commercial assays.
QuickFISH® assay
The QuickFISH® assays were performed on aliquots taken from positive blood culture bottles removed from the automated blood culture system and stored at 4 °C until sampling and Gram staining was performed, the same day or next morning. After the positive blood culture bottles had been given a code number, 10–15 droplets of the positive blood culture were transferred to an AdvanDx Filter Vial (OpGen, Gaithersburg, MD, United States). During the first substudy, Gram staining of positive blood cultures identifying Gram-positive cocci in cluster were selected for species identification with either the Staphylococcus and/or Enterococcus QuickFISH® assays (OpGen, United States). The second substudy focused on the positive blood cultures where Gram staining identified Gram-negative bacilli for further species identification using the Gram-negative QuickFISH® assay (OpGen, United States). The QuickFISH® assays were performed according to the manufacturer´s instructions, which also guided us to only analyse the first positive blood culture if more than one blood culture alerted (AdvanDx, 2014). The hybridized QuickFISH slides were immediately examined in a Leica DMR HC fluorescence microscope (Leica microsystems, Mannheim, Germany). The microscope had a 100X oil objective and was fitted with a dual-band filter (AC007 OpGen, United States). The QuickFISH slides include positive and negative controls that were read together with the clinical sample. The QuickFISH slides were read by eye independently by two different test operators.
Maldi sepsityper™ assay
During the first substudy, all positive blood cultures were analysed in parallel by both Maldi Sepsityper™ preparations methods, i.e. with or without extra formic acid treatment. The preparation of positive blood culture broths was performed according to the manufacturer’s instructions (Bruker Daltonics, Leipzig, Germany). The collected pellet was either put directly on to the MALDI target plate (Bruker Daltonics, Germany) or the pellet was further processed following the Bruker standard extraction procedure including the extra formic acid step, with some minor modification. The pellet was mixed with 300 μl deionized water and 900 μl 99.5% ethanol (Histolab, Gothenburg, Sweden). The suspension was vortexed for 1 min and centrifuged at 14.000 rpm for 2 min. The supernatant was discarded and centrifuged for an additional 2 min and air-dried. Sequentially, 5–50 μl each of 70% formic acid (Sigma, St Louis, MO) and 100% acetonitrile (Sigma, USA) was added to the pellet, and thoroughly mixed after each reagent was added. The sample was vortexed for 10 s and left standing for 2 min. The sample was centrifuged again at 14.000 rpm for another 2 min, and 1 μl of the supernatant was spotted onto the steel target plate. After both procedures definite species identification with MALDI-TOF MS was performed as described above.
Data analysis
The results obtained from the Maldi Sepsityper™ assay with or without formic acid treatment, were compared to those obtained by the reference method. The spectral scores were assessed and compared as identification rate using the spectral cut-off scores, according to the reference method. The identification rate was also assessed and compared at spectral cut-off score < 1.5. Species identified directly from monomicrobial blood cultures with the commercial assays were compared with the final species identification of these cultured monomicrobial blood cultures, by the reference method. The following results were considered as correctly identified microorganism during the assessment: (i) true positive, an organism identified by the commercial method that was identified by the reference method and reported to clinician as a final result, (ii) true negative, an organism not identified by the commercial method and neither identified by the reference method, (iii) false negative, an organism not identified by the commercial method but was identified by the reference method and reported to clinician as a final result, and lastly (iv) false positive, an organism identified by the commercial method but was not identified by the reference method. Concordance of identified microorganisms between the commercial assays and reference method was calculated as the number of true positives and true negatives divided by the total number of organisms identified. Sensitivity was calculated as the number of true positives divided by the number of true positives and false negatives, whereas specificity was calculated as the number of true negatives divided by the number of true negatives and false positives. For calculation of binomial proportion confidence intervals, the Clopper-Pearson method was applied using the PropCIs package for R version 3.2.3 (R Foundation for Statistical Computing, Vienna, Austria).
Ethical statement
This study is a clinical laboratory benchmarking of two commercial kits. The study does not involve the collection or reporting of patient data, and no patient intervention occurred with the obtained results.
Results
Benchmarking of the maldi sepsityper™ kit with or without extra formic acid treatment
Identification rate
The reference method identified microorganism at score >2.0 in 89.1% (173/194) of the cultured bacterial isolates (), whereof the most commonly identified microorganisms were CoNS, E. coli and S. aureus. Using the Maldi Sepsityper™ kit with or without the formic acid treatment, fewer microorganisms were identified at a score >2.0 (). Indeed, the extra formic acid treatment increased the identification rate at score >2.0 from 34.6% (62/179) to 60.1% (109/179), with a major impact on the identification rate of Gram-positives from 4.5% (8/179) to 31.8% (57/179) (). ‘No reliable identification’ as a result was observed for 2.1% (4/194) of the cultured bacterial isolates using the reference method compared to 45.3% (81/179) and 22.3% (40/179) of the positive blood cultures using the Maldi Sepsityper™ kit without formic acid treatment and Maldi Sepsityper™ with formic acid treatment, respectively (). If a cut-off score at 1.5 would have been used for species identification, the identification rate would have increased to 65.9% (118/179) for Maldi Sepsityper™ kit without formic acid treatment and 86.8% (154/179) for the Maldi Sepsityper™ with formic acid treatment.
Table 1. Identification rate using reference method on cultured isolates and Maldi Sepsityper™ assay with or without formic acid on positive blood culture.
Performance against routine diagnostic
During the first substudy, routine diagnostics recognised 100 positive blood cultures to be further cultured on solid media for subsequent analysis using the reference method, which resulted in 92 monomicrobial cultures and seven mixed cultures, which were reported as polymicrobial cultures as final results in clinical routine (). Among these seven positive blood cultures, the Maldi Sepsityper™ assay identified one microorganism or no identification was obtained at all (). In addition, from one patient, the reference method identified C. albicans at spectral score 1.8, being reported as yeast infection as a final result in clinical routine. The fungal growth was not identified by the Maldi Sepsityper™ assay (). Among the remaining 92 monomicrobial positive blood cultures being reported as final results in clinical routine, the reference method identified 63 Gram-positives and 25 Gram-negatives. Four cultured isolates had a score <1.7, reported as ‘no reliable identification’, which were neither identified by the Maldi Sepsityper™ kit. Altogether, Maldi Sepsityper™ kit without formic acid treatment showed a sensitivity of 51.1% (95% CI 40.2–61.9) which increased to 77.3% (95% CI 67.1–85.5) when applying the formic acid treatment, while the specificity decreased from 100.0% (95% CI 39.8–100.0) to 75.0% (95% CI 19.4–99.4), respectively (). The performance was considerably affected by the characteristics of the causative agents (). We observed 53.2% (95% CI 42.6–63.7) concordance of all identified species between the Maldi Sepsityper™ kit without formic acid treatment and the reference method and 77.2% (95% CI 67.2–85.3) concordance of all identified species between the Maldi Sepsityper™ with formic acid treatment and the reference method. The Maldi Sepsityper™ assay reported species identification after 0.5–1 h ().
Table 2. Polymicrobial and fungal cultures reported as final results in clinical routine during the first substudy.
Table 3. Performance of Maldi Sepsityper™ against routine diagnostics on samples from monomicrobial positive blood cultures.
Benchmarking of the QuickFISH® assay
Performance against routine diagnostic
The first substudy assessed the Staphylococcus and/or Enterococcus QuickFISH® in comparison with the reference method. Gram staining of the 179 positive blood cultures identified 57 Gram-positive cocci in cluster which were further analysed with either the Staphylococcus (n = 40) or the Enterococcus (n = 12) or both (n = 5) QuickFISH® kits. The reference method identified the majority of these monomicrobial cultures as 31.6% (18/57) CoNS, 28.0% (16/57) S. aureus followed by different streptococcal species 22.8% (13/57). The Staphylococcus QuickFISH® assay identified 42% (19/45) CoNS and 33.3% (15/45) S. aureus with 100.0% (95% CI 89.7–100.0) sensitivity and 100.0% (95% CI 71.5–100.0) specificity (). The only E. faecalis reported in clinical routine was correctly identified by the Enterococcus QuickFISH® kit and 93.8% (95% CI 69.8–99.8) were identified as true negatives ().
Table 4. Performance of the Staphylococcus QuickFISH® assay against routine diagnostics on samples from monomicrobial positive blood cultures.
Table 5. Performance of the Enterococcus QuickFISH® assay against routine diagnostics on samples from monomicrobial positive blood cultures.
During the second substudy, the Gram-negative QuickFISH® assay was assessed against the reference method. Gram staining of the 203 positive blood cultures identified 37 positive blood cultures containing Gram-negative bacilli, which were further selected for analysis by the Gram-negative QuickFISH® assay. The plating of these 37 positive blood cultures resulted in two mixed cultures on solid plates. These cultures were reported as polymicrobial cultures as a final result in clinical routine, the first K. oxytoca and E. coli, and the second E. faecalis and E. coli. The reference method identified the majority, 74.2% (26/35), of the monomicrobial positive blood cultures as E. coli. The QuickFISH® assay identified 84.6% (22/26) E. coli, 100% (1/1) P. aeruginosa and 100% (2/2) K. pneumonia as true positives, and 100.0% (6/6) as true negatives ().
Table 6. Performance of the Gram negative QuickFISH® assay against routine diagnostics on samples from monomicrobial positive blood cultures.
Regarding the polymicrobial cultures, the Gram-Negative QuickFISH® assay correctly identified the bacteria covered by the Gram-Negative QuickFISH® assay in both positive blood cultures. The Gram-Negative QuickFISH® assay showed a sensitivity of 86.2% (95% CI 68.3-96.1) and a specificity of 100.0% (95% CI 54.1–100.0). In all, the concordance of identified species between the QuickFISH assays and the reference method was 94.8% (95% CI 88.4-98.3), 98.4% (95% CI 91.3–100.0) for the Staphylococcus and Enterococcus QuickFISH® kit together, and 88.6% (95% CI 73.3–96.8) for the Gram-Negative QuickFISH®. The QuickFISH® assay could report species identification after 1 h ().
Discussion
Current composition of blood culture medium has facilitated analysis directly on samples from the positive blood culture bottles allowing earlier identification of the causative agent in bloodstream infections. In this study, we aimed to assess the performance of two different commercial blood culture identifications methods, the QuickFISH® and Maldi Sepsityper™ assays, by comparing each commercial assay with routine analysis of bloodstream infections at a clinical microbiology laboratory. We assessed the degree of concordance of identified microorganisms between the commercial assays and the reference method, turnaround time and identification rate during two substudies. Our results indicate that the use of the commercial assays may shorten the time to identification of causative agents in bloodstream infections. Nevertheless, the performance of the commercial assays was considerable affected by the characteristics of the causative agents.
At score >2.0, the reference method showed a satisfactory identification rate, identifying 92.2% (179/194) of the cultured isolates [Citation20] compared to the rather low identification rate 34.6% (62/179) and 60.1% (109/179) using the Maldi Sepsityper™ kit without formic acid treatment or with formic acid treatment respectively (). Previous studies reported higher identification rates ranging from 67–100% [Citation21–31] when evaluating the Maldi Sepsityper™ assay. However, some of these studies applied lower cut-off scores for definite species identification. Cut-off scores at 1.7 [Citation21,Citation26–28,Citation31], 1.6 [Citation24,Citation29] and even 1.5 [Citation19] have been used. If a cut-off score at 1.5 would have been used for definite species identification, the identification rate would have increased to 65.9% (118/179) for Maldi Sepsityper™ kit without formic acid treatment and 86.8% (154/179) for the Maldi Sepsityper™ with formic acid treatment. One possible explanation to our results is the challenge of correct identification of the Gram-positive bacteria ( and ), which is in agreement with a recent meta-analysis of the Maldi Sepsityper™ application showing that the test generally performs better in Gram-negatives than in Gram-positives [Citation32]. This preference had an impact on the performance of the Maldi Sepsityper™ kit in our study, as the reference method identified the majority of the monomicrobial cultures as Gram-positives, 68.5% (63/92), which follows the same trend that has been observed earlier in Swedish studies [Citation33]. The Maldi Sepsityper™ kit identified only 38.1% (24/63) of the Gram positives as true positives while the Maldi Sepsityper™ followed by formic acid treatment increased the true positives to 77.8% (49/63). The moderate concordance of the species identification between the Maldi Sepsityper™ and the reference method follows these results. In all, the Maldi Sepsityper™ assay was superior at identifying Gram-negative to Gram-positive bacteria ().
The QuickFISH® assay showed no such preferences, rather the performance was dependent on whether the causative agent was covered by the assay. The overall high concordance between the QuickFISH® assay and the reference method is consistent with other studies [Citation9–13], suggesting that QuickFISH® can provide a platform for accurate identification of bacterial species directly from monomicrobial blood cultures. The sensitivities for both Staphylococcus and Enterococcus QuickFISH® kit were satisfying ( and ), although the rather small numbers of positive blood cultures analysed by the Enterococcus QuickFISH® is a limitation during this study. However, among the positive blood cultures reported as final results in clinical routine, the majority were reported as CoNS and S. aureus. The Staphylococcus QuickFISH® was able to correctly differentiate CoNS from S. aureus with 100% sensitivity and 100% specificity. The Gram-Negative QuickFISH® was previously reported to achieve 95.7% concordance with routine methods using a combination of MALDI-TOF MS, standard biochemical tests, API 20E and Vitek 2 [Citation12]. We observed a slightly lower concordance of 88.6% (95% CI 73.3–96.8), probably due to subjective misinterpretations in the beginning of the Gram-Negative QuickFISH® evaluation (). Another limitation of our study was that the majority of positive blood cultures tested with the Gram-Negative QuickFISH® were positive for E. coli, thus we did not have the opportunity to fully assess the performance of the detection of other Gram-negatives.
Even though no clear-cut answer has been provided on whether the prognosis of polymicrobial infections is worse than that of monomicrobial infections [Citation34], it can be observed that the identification of polymicrobial cultures was not possible using the Maldi Sepsityper™ assay, as well as the challenge of identifying fungal cultures, all in accordance with other studies [Citation21,Citation25,Citation32]. A recent publication assessed an updated version of the Sepsityper module software able to detect mixed blood cultures through MALDI-TOF MS identification directly from a positive blood culture using the MALDI Sepsityper kit (Bruker Daltonik, Bremen, Germany). From 143 polymicrobial tested blood culture bottles, 34.3% (49/143) were completely identified by the module [Citation35].
Since QuickFISH® assays are capable of detecting up to a maximum of three different species per assay, the assay is limiting the identification of all microorganisms in polymicrobial positive blood cultures, which has been observed earlier [Citation12].
Early recognition of the causative agent represents an important factor influencing patient treatment and recovery [Citation36]. Thus, the turnaround time is important to consider when assessing for identification of bloodstream pathogens. However, the time needed for species identification by conventional culture-based methods differs between clinical laboratories depending on routines. Though turnaround times were estimated and not precisely measured, our results point towards that the use of the commercial assays may shorten the time to identification of causative agents in bloodstream infections. The QuickFISH® assay lead to a short turnaround time for pathogen identification, 1 h (), which has also been reported earlier, including only five minutes hands-on time [Citation9–11,Citation13]. The Maldi Sepsityper™ kit can be timesaving as well, but it needs more manual hands-on time. Furthermore, since the two commercial assays are used directly on positive blood cultures, the clinician could get the final species identification the same day as blood was withdrawn from the patient.
Currently, it is not possible to use these commercial assays for antibiotic susceptibility testing in routine workflow, and therefore, culture of pure isolates is still needed for this purpose. However, there has been efforts to address this problem with MALDI-TOF MS applications detecting bacterial proteins involved in antibiotic resistance, such as the β-Lactamase enzyme [Citation18,Citation37].
Previous European studies focusing on direct costs per sepsis patient have yielded an estimated range from 23,000–42,000 EUR [Citation2,Citation38,Citation39]. The major determinant of the direct costs is the length of stay in the hospital, clearly illustrating the potential savings in shortening the time to correct diagnosis of patients with bloodstream infection. A recent review has shown that the use of rapid diagnostic tests such as the PNA-FISH is a cost-effective strategy that was associated with high therapeutic effectiveness and health care cost savings [Citation40]. In addition, it has been demonstrated that direct identification of organism in positive blood cultures bottles using MALDI-TOF MS or QuickFISH® may have a clinical impact in patients with bacteraemia and improve appropriateness of antibiotic therapy [Citation10,Citation41–43].
Conclusion
The performance data of the assessed methods suggest that the use of a commercial assay may indeed be a good addition to the current routine laboratory diagnostics for an earlier but still sufficiently accurate identification of microorganisms directly from positive blood culture bottles. Based on the result from this study, we suggest a workflow where the current Gram stain could guide the choice of downstream species identification methodology. The polymicrobial –, as well as the fungal –, blood cultures should still be analysed only by the reference method. Considering monomicrobial Gram-negative cultures, the Maldi Sepsityper™ assay may well deliver the correct species identification earlier than the reference method. As Gram-positive cocci in cluster is currently the most commonly encountered bacterial morphotype in positive blood cultures [Citation33], the Staphylococcus QuickFISH® can serve as a complement to the reference method, enable the laboratory to accurately differentiating S. aureus from CoNS in blood cultures in 1 h.
Conflicts of interest
The authors declare that they have no conflict of interest regarding this article.
Acknowledgments
▪
Additional information
Funding
References
- Adhikari NK, Fowler RA, Bhagwanjee S, et al. Critical care and the global burden of critical illness in adults. Lancet. 2010;376:1339–1346.
- Alvarez J, Mar J, Varela-Ledo E, et al. Cost analysis of real-time polymerase chain reaction microbiological diagnosis in patients with septic shock. Anaesth Intensive Care. 2012; 40:958–963.
- Ziegler R, Johnscher I, Martus P, et al. Controlled clinical laboratory comparison of two supplemented aerobic and anaerobic media used in automated blood culture systems to detect bloodstream infections. J Clin Microbiol. 1998;36:657–661.
- Ljungstrom L, Enroth H, Claesson BE, et al. Clinical evaluation of commercial nucleic acid amplification tests in patients with suspected sepsis. BMC Infect Dis 2015;15.
- Oliveira K, Procop GW, Wilson D, et al. Rapid identification of Staphylococcus aureus directly from blood cultures by fluorescence in situ hybridization with peptide nucleic acid probes. J Clin Microbiol. 2002;40:247–251.
- Perry-O'Keefe H, Rigby S, Oliveira K, et al. Identification of indicator microorganisms using a standardized PNA FISH method. J Microbiol Methods. 2001;47:281–292.
- Stender H. PNA FISH: an intelligent stain for rapid diagnosis of infectious diseases. Expert Rev Mol Diagn. 2003;3:649–655.
- Frickmann H, Zautner AE, Moter A, et al. Fluorescence in situ hybridization (FISH) in the microbiological diagnostic routine laboratory: a review. Crit Rev Microbiol. 2017;43:263–293.
- Carretto E, Bardaro M, Russello G, et al. Comparison of the Staphylococcus QuickFISH BC test with the tube coagulase test performed on positive blood cultures for evaluation and application in a clinical routine setting. J Clin Microbiol. 2013;51:131–135.
- Koncelik DL, Hernandez J. The impact of implementation of rapid QuickFISH testing for detection of coagulase-negative Staphylococci at a community-based hospital. Am J Clin Pathol. 2016;145:69–74.
- Deck MK, Anderson ES, Buckner RJ, et al. Multicenter evaluation of the Staphylococcus QuickFISH method for simultaneous identification of Staphylococcus aureus and coagulase-negative staphylococci directly from blood culture bottles in less than 30 minutes. J Clin Microbiol. 2012;50:1994–1998.
- Martinez RM, Bauerle ER, Fang FC, et al. Evaluation of three rapid diagnostic methods for direct identification of microorganisms in positive blood cultures. J Clin Microbiol. 2014;52:2521–2529.
- Deck MK, Anderson ES, Buckner RJ, et al. Rapid detection of Enterococcus spp. direct from blood culture bottles using Enterococcus QuickFISH method: a multicenter investigation. Diagn Microbiol Infect Dis. 2014;78:338–342.
- Savini V, Gherardi G, Marrollo R, et al. Could beta-hemolytic, group B Enterococcus faecalis be mistaken for Streptococcus agalactiae?. Diagn Microbiol Infect Dis. 2015;82:32–33.
- Abdelhamed AM, Zhang SX, Watkins T, et al. Multicenter evaluation of Candida QuickFISH BC for identification of Candida species directly from blood culture bottles. J Clin Microbiol. 2015;53:1672–1676.
- Seng P, Drancourt M, Gouriet F, et al. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. 2009;49:543–551.
- Fenselau C, Demirev PA. Characterization of intact microorganisms by MALDI mass spectrometry. Mass Spectrom Rev. 2001;20:157–171.
- Dubourg G, Lamy B, Ruimy R. Rapid phenotypic methods to improve the diagnosis of bacterial bloodstream infections: meeting the challenge to reduce the time to result. Clin Microbiol Infect 2018..
- Schubert S, Weinert K, Wagner C, et al. Novel, improved sample preparation for rapid, direct identification from positive blood cultures using matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry. J Mol Diagn. 2011; 3:701–706.
- Bizzini A, Greub G. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry, a revolution in clinical microbial identification. Clin Microbiol Infect. 2010;16:1614–1619.
- Buchan BW, Riebe KM, Ledeboer NA. Comparison of the MALDI Biotyper system using Sepsityper specimen processing to routine microbiological methods for identification of bacteria from positive blood culture bottles. J Clin Microbiol. 2012;50:346–352.
- Chen JH, Ho PL, Kwan GS, et al. Direct bacterial identification in positive blood cultures by use of two commercial matrix-assisted laser desorption ionization-time of flight mass spectrometry systems. J Clin Microbiol. 2013;51:1733–1739.
- Loonen AJ, Jansz AR, Stalpers J, et al. An evaluation of three processing methods and the effect of reduced culture times for faster direct identification of pathogens from BacT/ALERT blood cultures by MALDI-TOF MS. Eur J Clin Microbiol Infect Dis. 2012;31:1575–1583.
- Martiny D, Dediste A, Vandenberg O. Comparison of an in-house method and the commercial Sepsityper kit for bacterial identification directly from positive blood culture broths by matrix-assisted laser desorption-ionisation time-of-flight mass spectrometry. Eur J Clin Microbiol Infect Dis. 2012;31:2269–2281.
- Kok J, Thomas LC, Olma T, et al. Identification of bacteria in blood culture broths using matrix-assisted laser desorption-ionization Sepsityper and time of flight mass spectrometry. PLoS One. 2011;6:e23285.
- Lagace-Wiens PR, Adam HJ, Karlowsky JA, et al. Identification of blood culture isolates directly from positive blood cultures by use of matrix-assisted laser desorption ionization-time of flight mass spectrometry and a commercial extraction system: analysis of performance, cost, and turnaround time. J. Clin Microbiol. 2012;50:3324–3328.
- Klein S, Zimmermann S, Kohler C, et al. Integration of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in blood culture diagnostics: a fast and effective approach. J Med Microbiol. 2012;61:323–331.
- Saffert RT, Cunningham SA, Mandrekar J, et al. Comparison of three preparatory methods for detection of bacteremia by MALDI-TOF mass spectrometry. Diagn Microbiol Infect Dis. 2012;73:21–26.
- Schieffer KM, Tan KE, Stamper PD, et al. Multicenter evaluation of the Sepsityper extraction kit and MALDI-TOF MS for direct identification of positive blood culture isolates using the BD BACTEC FX and VersaTREK((R)) diagnostic blood culture systems. J Appl Microbiol. 2014;116:934–941.
- Yonetani S, Ohnishi H, Ohkusu K, et al. Direct identification of microorganisms from positive blood cultures by MALDI-TOF MS using an in-house saponin method. Int J Infect Dis. 2016;52:37–42.
- Tanner H, Evans JT, Gossain S, et al. Evaluation of three sample preparation methods for the direct identification of bacteria in positive blood cultures by MALDI-TOF. BMC Res Notes. 201710:48.
- Morgenthaler NG, Kostrzewa M. Rapid identification of pathogens in positive blood culture of patients with sepsis: review and meta-analysis of the performance of the sepsityper kit. Int J Microbiol. 2015;2015:1.
- Holmbom M, Giske CG, Fredrikson M, et al. 14-Year survey in a Swedish county reveals a pronounced increase in bloodstream infections (BSI). Comorbidity - an independent risk factor for both BSI and mortality. PLoS One. 2016;11:e0166527.
- Pavlaki M, Poulakou G, Drimousis P, et al. Polymicrobial bloodstream infections: Epidemiology and impact on mortality. J Glob Antimicrob Resist. 2013;1:207–212.
- Scohy A, Noel A, Boeras A, et al. Evaluation of the Bruker(R) MBT Sepsityper IVD module for the identification of polymicrobial blood cultures with MALDI-TOF MS. Eur J Clin Microbiol Infect Dis. 2018.
- Retamar P, Portillo MM, Lopez-Prieto MD, et al. Impact of inadequate empirical therapy on the mortality of patients with bloodstream infections: a propensity score-based analysis. Antimicrob Agents Chemother. 2012;56:472–478.
- Hrabak J, Chudackova E, Walkova R. Matrix-assisted laser desorption ionization-time of flight (maldi-tof) mass spectrometry for detection of antibiotic resistance mechanisms: from research to routine diagnosis. Clin Microbiol Rev. 2013;26:103–114.
- Chalupka AN, Talmor D. The economics of sepsis. Crit Care Clin. 2012;28:57–76.
- Burchardi H, Schneider H. Economic aspects of severe sepsis: a review of intensive care unit costs, cost of illness and cost effectiveness of therapy. Pharmacoeconomics. 2004;22:793–813.
- Pliakos EE, Andreatos N, Shehadeh F, et al. The cost-effectiveness of rapid diagnostic testing for the diagnosis of bloodstream infections with or without antimicrobial stewardship. Clin Microbiol Rev. 2018; 31.
- Almangour TA, Alhifany AA, Tabb DE. Development and validation of a decision-making stratification algorithm to optimize the use of rapid diagnostic testing for patients with staphylococcus bacteremia. Can J Infect Dis Med Microbiol. 2017;2017:1.
- Vlek AL, Bonten MJ, Boel CH. Direct matrix-assisted laser desorption ionization time-of-flight mass spectrometry improves appropriateness of antibiotic treatment of bacteremia. PLoS One. 2012;7:e32589.
- French K, Evans J, Tanner H, et al. The clinical impact of rapid, direct MALDI-ToF identification of bacteria from positive blood cultures. PLoS One. 2016;11:e0169332.
