Abstract
Background
Bacterial infections complicating COVID-19 are rare but present a challenging clinical entity. The aim of this study was to evaluate the incidence, aetiology and outcome of severe laboratory-verified bacterial infections in hospitalised patients with COVID-19.
Methods
All laboratory-confirmed patients with COVID-19 admitted to specialised healthcare hospitals in the Capital Province of Finland during the first wave of COVID-19 between 27 February and 21 June 2020 were retrospectively studied. We gathered the blood and respiratory tract culture reports of these patients and analysed their association with 90-day case-fatality using multivariable regression analysis.
Results
A severe bacterial infection was diagnosed in 40/585 (6.8%) patients with COVID-19. The range of bacteria was diverse, and the most common bacterial findings in respiratory samples were gram-negative, and in blood cultures gram-positive bacteria. Patients with severe bacterial infection had longer hospital stay (mean 31; SD 20 days) compared to patients without (mean 9; SD 9 days; p < 0.001). Case-fatality was higher with bacterial infection (15% vs 11%), but the difference was not statistically significant (OR 1.38 CI95% 0.56–3.41).
Conclusions
Severe bacterial infection complicating COVID-19 was a rare occurrence in our cohort. Our results are in line with the current understanding that antibiotic treatment for hospitalised COVID-19 patients should only be reserved for situations where a bacterial infection is strongly suspected. The ever-evolving landscape of the pandemic and recent advances in immunomodulatory treatment of COVID-19 patients underline the need for continuous vigilance concerning the possibility and frequency of nosocomial bacterial infections.
Introduction
Coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has a broad spectrum of manifestations affecting the upper respiratory tract and lungs, blood coagulation, heart, gastrointestinal system, and nervous system among other organs [Citation1]. Secondary bacterial infections are common complications in influenza [Citation2] or other viral respiratory infections, but rare in patients with COVID-19, a large meta-analysis estimating the incidence between 10 and 20% [Citation3]. Several different mechanisms including airway epithelium damage, abnormal inflammatory reactions, and dysregulation of innate and acquired immune responses may contribute to the development of bacterial infections in patients with viral respiratory disease. These mechanisms are observed also in COVID-19 which makes the low level of secondary bacterial infections in these patients somewhat surprising [Citation1]. Both pulmonary imaging findings and laboratory inflammatory parameters caused by COVID-19 may be difficult to distinguish from potential bacterial complications, which commonly results in antibiotic treatment and concern for the overuse of antibiotics, especially during the first wave, was raised globally [Citation4]. Furthermore, as a novel pathogen emerges, initial responses and treatment guidelines are based on very limited information on the frequency of serious sequelae including community-acquired and nosocomial bacterial infections.
The aim of this study was to define the incidence and describe the microbial aetiology of severe community-acquired and nosocomial bacterial infections among all hospitalised patients with COVID-19 in the Capital Province of Finland by using a population-based quality registry together with laboratory registry data during the first wave of COVID-19 in the spring of 2020.
Materials and methods
This is a retrospective observational study utilising the population-based quality registry of hospitalised COVID-19 patients in the hospital district of Helsinki and Uusimaa, the Capital Province of Finland. The hospital district of the Capital Province of Finland (HUS Helsinki University Hospital) provides specialised healthcare to 1.7 million inhabitants with approximately 3000 beds and 2.7 million annual patient visits and is one of the largest healthcare organisations in Europe. Inpatient care of COVID-19 patients in Finland is organised through publicly funded healthcare and hospitals, so private healthcare was not included in the quality registry [Citation5]. Nursing home residents and residents in assisted living are generally treated in primary care hospitals, which are also excluded from the quality registry.
Ethical approval
The study was institutionally approved (HUS/157/2020 and HUS/41/2021) as an observational registry study and patient consent was waived, allowing the inclusion of all consecutive patients.
The patient inclusion criteria, demographics, clinical characteristics, and outcomes have been described previously [Citation5]. Extensive demographic and clinical details, such as length of stay and mortality of the hospitalised patients were collected in the quality registry. In this study, in addition to the original quality registry data, the complete duration of antibiotic treatment of each patient was recorded and laboratory registry data were collected for microbiological findings in categories listed in Supplement 1.
All microbiological samples collected during hospitalisation were analysed in HUSLAB, HUS Diagnostic Centre, Helsinki. Methods, reagents and laboratory equipment used for routine tests are summarised in Supplement 2. Reverse transcription polymerase chain reaction (RT-PCR) tests used for SARS-CoV-2 detection in clinical samples in our laboratory have been described previously [Citation6,Citation7].
Severe microbiologically confirmed bacterial infection was defined as either bloodstream infection with a finding in blood culture or bacterial pneumonia with a significant finding in respiratory tract bacterial culture or a combination of both. Significant respiratory tract bacterial culture is defined further below. Both community-acquired bacterial infections (positive culture collected ≤48 h after hospital admission) and nosocomial bacterial infections (positive culture collected >48 h after hospital admission) were included in the analysis.
Blood culture
One blood culture set consisted of 4 bottles: 2 aerobic and 2 anaerobic bottles. All blood culture findings were considered significant, except coagulase-negative staphylococcus detected in a single bottle. In cases of typical skin contaminants recovered from >1 bottles, the patient records were evaluated by an infectious diseases specialist to determine the clinical significance. Antibiotic susceptibility testing was performed for all recovered isolates using EUCAST methodology (www.eucast.org). Blood cultures were collected when the treating physician deemed necessary, no specific protocol was in place.
Respiratory tract bacterial culture
Sample types included in the evaluation of possible bacterial pneumonia were tracheal aspirates, bronchoalveolar lavage, sputum, pleural fluid, and samples collected from a tracheostomy tube.
Species-level identification and antimicrobial susceptibility testing results were reported for findings that were considered potential pathogens (e.g. Staphylococcus aureus, Streptococcus pneumoniae, gram-negative bacilli). Species-level identification for yeasts recovered in bacterial culture samples was not routinely performed or reported but a comment was added to recommend fungal culture in case of suspicion of fungal infection. When the laboratory reported growth of “mixed aerobic flora” a comment describing the most abundantly present bacteria was added. No antimicrobial susceptibility testing was performed in such cases or in cases where the growth was considered to represent commensal microbial flora.
Statistical analyses
Categorical variables were compared with Pearson’s X2 test. Non-categorical variables were tested for normal distribution using the Shapiro–Wilk test and reported as mean with standard deviation or median with interquartile range, as appropriate. However, hospital and intensive care unit (ICU) length of stay were reported using both. Odds ratios (OR) with 95% confidence intervals (CI) were calculated. We did not consider variables with missing data (BMI, smoking, alcohol, or illegal drugs) for inclusion in the multivariable model. Tests were two-tailed and p< .05 was considered significant. Analyses were done with SPSS 25.0.0 IBM SPSS Statistics for Windows, Version 25.0 (IBM Corp., Armonk, NY).
Results
Patient demographics
During the first wave of the pandemic in Finland, between February 27th and June 21st, 2020, 5471 people were diagnosed with SARS-CoV2 infection in the study region and 585 patients with COVID-19 were admitted to specialised healthcare hospitals of the Capital Province of Finland. Of these patients, 132 (22.6%) were treated in ICU. A severe bacterial infection was diagnosed in 40/585 (6.8%) patients (). Most of the patients with severe infection (n = 35, 87.5%) were treated in the ICU, while only five (12.5%) patients were treated in a standard ward throughout the admission. Patient demographics, risk behaviour and conditions are presented in . Male sex, sleep apnoea, hypertension and diabetes were overrepresented in the group that had severe bacterial infections. The age range in our cohort was 4 months to 95 years while the age range of patients with severe bacterial infection was 35 to 89 years.
Figure 1. Flow chart for distribution of intensive care unit (ICU) treatment need and severe bacterial infections among hospitalised COVID-19 patients during the first wave of the COVID-19 pandemic in the Capital Province of Finland.
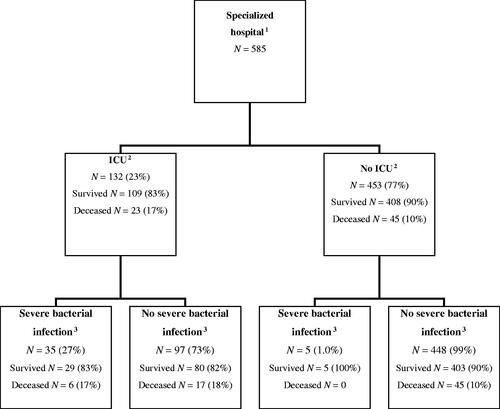
Table 1. Patient demographics, risk behaviour and conditions of 585 patients with laboratory confirmed COVID-19 and treated in specialised healthcare hospital stratified according to the occurrence of severe bacterial infections (bacteremic or culture verified respiratory tract infections).
The incidence and microbial aetiology of severe bacterial infections
A total of 742 blood culture sets were collected from 483 (83%) patients, 128/132 (97%) of ICU patients and 355/453 (78%) of non-ICU patients: 15 patients had one or more positive blood cultures (). A significant bloodstream infection was diagnosed in twelve patients and three patients were deemed to have contaminant growth. Over half (7/12) of the bloodstream infections were diagnosed in the ICU group. Overall, 2.5% (12/483) of patients had positive blood cultures and the frequency in patients treated outside the ICU was 1.4% (5/355) and for ICU patients 5.5% (7/128). Supplement 3 contains a summary of the cases with significant blood culture findings.
Table 2. Aetiology of severe bacterial infections: blood stream infections (BSI) and bacterial pneumonias.
Respiratory samples for bacterial culture were collected from 94 patients and 90 of them were treated in the ICU. Number of different sample types included in the evaluation of possible bacterial pneumonia were tracheal aspirates (n = 224), bronchoalveolar lavage (n = 16), sputum (n = 1), pleural fluid (n = 5) and samples collected from a tracheostomy tube (n = 3). Significant growth was detected in the samples of 33 patients, all from the ICU (). Five patients had the same finding in the respiratory tract culture and in the blood culture.
In our patient cohort, a total of 40/585 (6.8%) patients had a severe bacterial infection. Five patients had both a bloodstream infection and a microbiologically confirmed bacterial infection of the respiratory tract, whereas seven patients had only a bloodstream infection, and 28 patients had only a finding in the respiratory tract culture. Most samples (31 patients) with significant growth were from nosocomial infections. Nine patients had a community-acquired bacterial infection, six of whom had positive blood cultures and three had significant growth in respiratory tract cultures.
The most common microbes found in respiratory tract cultures were S. aureus, Pseudomonas aeruginosa and Escherichia coli. Strains producing extended spectrum betalactamase (ESBL) were recovered from five patients (four E. coli strains and one K. pneumoniae strain; all from respiratory tract cultures). No carbapenem-resistant strains were identified. S. aureus was the most common finding in blood culture (3 patients). No MRSA was found. The significant bacterial culture findings are summarised in . One Aspergillus fumigatus was recovered in bacterial culture but was considered colonisation.
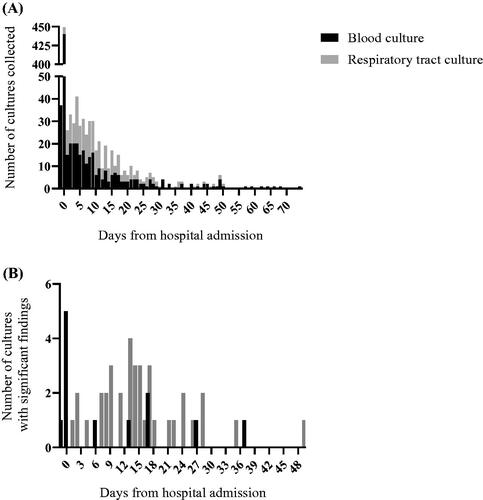
The timing of blood culture and respiratory tract sample collection and the timing of significant findings are depicted in , respectively. Most of the microbiological sampling was done early during hospitalisation: 54% of all respiratory tract and blood culture samples were collected <48 h after hospitalisation, but the proportion of samples with significant growth increased over time.
Figure 2. Timing of microbiological sampling and significant blood culture and respiratory tract findings. Most samples were collected early during hospitalisation (A), but significant findings show an increasing trend towards longer hospitalisation (B). Significant findings reach a second peak at two weeks after hospital admission, while the first peak appears at the time of hospital admission when the majority of all blood cultures were collected. The X-axis in (A) and (B) shows time in days from hospital admission. aSpecialised hospital healthcare (including Helsinki University Hospital). bIntensive care unit treatment. cBlood culture positive infection and/or respiratory tract bacterial culture positive.
Patient outcomes
Patients with a severe bacterial infection had a 28-day case-fatality rate similar to that of the patients who did not have a severe bacterial infection: 13% vs 10% (). At 90 days the difference grew to 15% vs 11%, (OR 1.38 CI95% 0.56–3.41) but did not reach statistical significance. Patients with severe bacterial infection had longer hospital stays (mean 31.4 vs 8.5 days) and longer ICU treatments (22.9 vs 11 days) as compared to those without. The time from COVID-19 diagnosis to hospital admission was shorter for patients who had a severe bacterial infection (0.8 vs 2.7 days) but the time from symptom onset to COVID-19 diagnosis did not differ between the groups. The time from symptoms to death was longer for patients with severe bacterial infection (28.2 vs 17.5 days) (). In multivariable analysis parameters independently associated with severe bacterial infection were hypertension, obstructive sleep apnoea, acute pulmonary embolism and dialysis treatment (). However, when including only patients treated in ICU, only obstructive sleep apnoea and acute pulmonary embolism were independently associated with severe bacterial infection ().
Table 3. Clinical management, complications and outcome of 585 patients with laboratory confirmed COVID-19 in specialised healthcare hospital and stratified according to occurrence of severe bacterial infection (bacteremic or culture verified respiratory tract infections).
Table 4. Multivariable analysis for risk factors of severe bacterial infections (bacteremic or culture verified respiratory tract infections) in patients with laboratory confirmed COVID-19 disease treated in specialised healthcare hospital and categorised according to the whole patient cohort (N = 585) (a) and intensive care unit (N = 132) (b).
Use of antibiotics
In our patient cohort, antibiotic treatment was initiated for 522/585 (89%) patients. The recommended empiric antibiotic for hospitalised COVID-19 patients was ceftriaxone during the study period. Duration of antibiotic treatment in this group was 1-91 days, median 6 days (IQR 4–9). Severe bacterial infection was microbiologically confirmed in 40/522 (7.7%) patients receiving antimicrobial therapy.
Discussion
In our study, the frequency of severe bacterial infections, 40/585 (6.8%), seemed to be even lower than in previous studies [Citation8–11]. The association of severe bacterial infections as a complication on mortality was small and did not reach statistical significance. However, this might be due to the small sample size and the low frequency of severe bacterial infections. Blood cultures were drawn from 83% of patients which means that it is unlikely that we would have missed many bacteremic cases. Half of bloodstream infections were community-acquired also suggesting a low risk for nosocomial bloodstream infections in COVID-19 patients.
The number of significant findings in blood culture was extremely low in our patient material. In the current treatment protocols of COVID-19 dexamethasone for patients with the increasing need for supplemental oxygen and tocilizumab for certain ICU patients have an important role [Citation12,Citation13], which was not the case during the study period early in the pandemic. These new developments may predispose patients to bacterial infections through immunosuppression, so current evaluations on the incidence of bacterial infections in COVID-19 patients are continually needed [Citation14,Citation15]. Our study presents a baseline for the incidence in a population with low prevalence of resistant bacteria and can be used as a reference point for evaluating the safety of current treatment regimens in terms of increased risk for bacterial infections.
Previous studies have described a higher blood culture contaminant rate [Citation16,Citation17]. Hospital and ICU capacities were at the time only moderately strained in Finland compared to parts of the world that were struggling with massive COVID-19 surges, so ensuring adequate personnel to maintain good hospital hygiene practices and having highly specialised personnel for blood culture sampling was easier to accomplish in Finland. This has probably lowered the probability of skin contaminants growing in blood cultures.
Patients with severe bacterial infection had longer hospital stays and longer episodes in the ICU than patients who did not have a microbiologically confirmed bacterial infection. The time from symptoms to death was longer for patients with severe bacterial infection, which seems to point to the potential of COVID-19 to progress rapidly enough to result in fatal outcomes before the emergence of nosocomial infections.
Previous studies have found critically ill COVID-19 patients to be at an increased risk for nosocomial infection [Citation18]. Prolonged hospital treatment, especially in the ICU is, in itself, a risk factor for bacterial infections, especially ventilator-associated bacterial pneumonia and as we only took microbiologically verified pneumonias into account, we have certainly missed some cases, since even in evident cases of pneumonia bacterial cultures may be negative.
Rates of antimicrobial resistance were low as expected since in general the prevalence of resistant bacteria has remained low in Finland [Citation19]. This might be one of the key factors why those patients who had severe bacterial infections fared almost as well as those without since in populations with high resistance rates, severe bacterial infections seem to have led to high mortality [Citation20]. It is notable, that despite low rates of resistance in Finland, ESBL-producing E. coli strains outnumbered susceptible E. coli strains in the respiratory tract cultures (3 patients vs 2 patients) in our study, which could be explained by selection pressure due to empiric ceftriaxone treatment.
COVID-19-associated pulmonary aspergillosis appeared to be non-existent in this study population. Previous studies have found higher incidences, even as high as 30% in ICU patients receiving mechanical ventilation, though inter-study variability has been wide and differences in definitions and diagnostic criteria further complicate comparisons of incidence [Citation21,Citation22].
Unexpectedly, sleep apnoea emerged as an independent factor increasing the likelihood of severe bacterial infection among COVID-19 patients. The mechanism or pathophysiology behind this phenomenon is unclear and requires further investigation, even though the higher risk of hospitalisation in this patient group has been previously established [Citation23]. Epithelial damage, decreased ventilation and mechanical factors may play a role as well as some lifestyle factors that would predispose to sleep apnoea.
Pulmonary embolism was a more common complication in patients who also had a severe bacterial infection when compared with COVID-19 patients with no severe bacterial infection. This was the case both in ICU and non-ICU patients. The temporal and causal relation between these complications remains unclear since even though the date of diagnosis of bacterial infection and pulmonary embolism can be determined retrospectively through inspection of patient records, the date of the diagnostic CT scan does not necessarily reflect the actual date of the occurrence of the embolism. Similarly, the date of sampling does not necessarily accurately point out the beginning of the bacterial infection. Moreover: both diagnostic imaging and microbiological sampling are naturally done more extensively and actively in patients who are worsening, which may be attributed to a variety of reasons, which in turn leads to a more detailed acknowledgment of concomitant complications that may have contributed to the clinical deterioration to a varying extent.
As the pandemic has progressed, and especially in the era of universal vaccination programs, it is becoming increasingly common for a COVID-19-positive patient to be hospitalised for reasons unrelated to COVID-19. During our study period, spring 2020, this was still a rare occurrence and is unlikely to have had a large impact on our results, but it is possible that a few such patients are included in our cohort, and it is possible that the confirmed bacterial infection is in fact the primary reason for hospitalisation and positive SARS-CoV-2 RT-PCR is merely a coincidental finding.
Our results further emphasise the need to limit antibiotic treatment of COVID-19 patients to only those patients who present with signs and symptoms raising suspicion of bacterial infection since the incidence of severe bacterial infections was low [Citation17,Citation24,Citation25]. Routine empirical antibiotic treatment should thus be avoided when possible. Empirical antibiotic treatment has likely contributed to the low occurrence of severe bacterial infections in our cohort but may have negatively affected the proportion of infections caused by more resistant strains. The low overall occurrence of severe bacterial infections could aid clinicians in decision making although severe forms of COVID-19 can mimic bacterial infection. Further research is warranted to clarify the role of bacterial infections in COVID-19 in order to inform clinical decision-making and the use of antibiotic treatment.
In conclusion, despite extensive blood culture sampling, only 2.1% of COVID-19 patients had a bacteremic infection of which half were present already on hospital admission. Bacteremias were observed in patients with a clinical infection focus and they were mainly caused by gram-positive organisms. Culture verified bacterial pneumonia was mainly caused by gram-negative organisms and was observed later in the course of COVID-19 treatment. Severe bacterial infection was associated with prolonged hospital stay and ICU treatment.
Author contributions
MJA: Conceptualisation, investigation, data curation, visualisation, writing – original draft, EK: Conceptualisation, investigation, data curation, writing – original draft. EF: Conceptualisation, formal analysis, writing – review and editing. APS: Conceptualisation, writing – review and editing. NF: Conceptualisation, writing – review and editing AM: Conceptualisation, data curation, writing – review and editing. SMK: Conceptualisation, data curation, writing – review and editing. SKU: Conceptualisation, resources, writing – review and editing, supervision, project administration. ML: resources, writing – review and editing, supervision, project administration. AJ: Conceptualisation, resources, writing – review and editing, project administration. HJ: Conceptualisation, formal analysis, resources, writing – original draft, writing – review and editing, supervision, project administration.
Supplemental Material
Download Zip (79.4 KB)Disclosure statement
AJ reports lecture honoraria from Astellas, Gilead, GlaxoSmithKLine, Sanofi and ThermoFisher and consultation fees from Gilead, GlaxoSmithKline, Sanofi, Sobi and Roche outside the scope of the present work.
Additional information
Funding
References
- Thakur V, Ratho RK, Kumar P, et al. Multi-organ involvement in covid-19: beyond pulmonary manifestations. JCM. 2021;10(3):446.
- Bakaletz LO. Viral–bacterial co-infections in the respiratory tract. Curr Opin Microbiol. 2017;35:30–35.
- Langford BJ, So M, Raybardhan S, et al. Bacterial co-infection and secondary infection in patients with COVID-19: a living rapid review and meta-analysis. Clin Microbiol Infect. 2020;26(12):1622–1629.
- Langford BJ, So M, Raybardhan S, et al. Antibiotic prescribing in patients with COVID-19: rapid review and meta-analysis. Clin Microbiol Infect. 2021;27(4):520–531.
- Forsblom E, Silén S, Kortela E, et al. Male predominance in disease severity and mortality in a low covid-19 epidemic and low case-fatality area – a population-based registry study. Infect Dis (Lond). 2021;53(10):789–799.
- Jokela P, Jääskeläinen AE, Jarva H, et al. SARS-CoV-2 sample-to-answer nucleic acid testing in a tertiary care emergency department: evaluation and utility. J Clin Virol. 2020;131:104614.
- Mannonen L, Kallio-Kokko H, Loginov R, et al. Comparison of two commercial platforms and a Laboratory-Developed test for detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA. J Mol Diagn. 2021;23(4):407–416.
- Buetti N, Ruckly S, de Montmollin E, et al. COVID-19 increased the risk of ICU-acquired bloodstream infections: a case–cohort study from the multicentric OUTCOMEREA network. Intensive Care Med. 2021;47(2):180–187.
- Cohen R, Finn T, Babushkin F, et al. High rate of bacterial respiratory tract co-infections upon admission amongst moderate to severe COVID-19 patients. Infect Dis. 2022;54(2):134–144.
- Garcia-Vidal C, Sanjuan G, Moreno-García E, et al. Incidence of co-infections and superinfections in hospitalized patients with COVID-19: a retrospective cohort study. Clin Microbiol Infect. 2021;27(1):83–88.
- Ripa M, Galli L, Poli A, et al. Secondary infections in patients hospitalized with COVID-19: incidence and predictive factors. Clin Microbiol Infect. 2021;27(3):451–457.
- Abani O, Abbas A, Abbas F, et al. Tocilizumab in patients admitted to hospital with COVID-19 (RECOVERY): a randomised, controlled, open-label, platform trial. Lancet. 2021;397(10285):1637–1645.
- Horby P, Lim WS, Emberson JR, RECOVERY Collaborative Group, et al. Dexamethasone in hospitalized patients with covid-19. N Engl J Med. 2021;384(8):693–704.,
- Gragueb-Chatti I, Lopez A, Hamidi D, et al. Impact of dexamethasone on the incidence of ventilator-associated pneumonia and blood stream infections in COVID-19 patients requiring invasive mechanical ventilation: a multicenter retrospective study. Ann Intensive Care. 2021;11(1):87.
- Pettit NN, Nguyen CT, Mutlu GM, et al. Late onset infectious complications and safety of tocilizumab in the management of COVID-19. J Med Virol. 2021;93(3):1459–1464.
- Karami Z, Knoop BT, Dofferhoff ASM, et al. Few bacterial co-infections but frequent empiric antibiotic use in the early phase of hospitalized patients with COVID-19: results from a multicentre retrospective cohort study in The Netherlands. Infect Dis. 2021;53(2):102–110.
- Hughes S, Troise O, Donaldson H, et al. Bacterial and fungal coinfection among hospitalized patients with COVID-19: a retrospective cohort study in a UK secondary-care setting. Clin Microbiol Infect. 2020;26(10):1395–1399.
- Grasselli G, Scaravilli V, Mangioni D, et al. Hospital-acquired infections in critically ill patients with COVID-19. Chest. 2021;160(2):454–465.
- Surveillance of Antimicrobial Resistance in Europe. 2020. Data. Executive Summary
- Li J, Wang J, Yang Y, et al. Etiology and antimicrobial resistance of secondary bacterial infections in patients hospitalized with COVID-19 in Wuhan, China: a retrospective analysis. Antimicrob Resist Infect Control. 2020;9(1):1–7.
- Alanio A, Dellière S, Fodil S, et al. Prevalence of putative invasive pulmonary aspergillosis in critically ill patients with COVID-19. Lancet Respir Med. 2020;8(6):e48–e49.
- Bartoletti M, Pascale R, Cricca M, et al. Epidemiology of invasive pulmonary aspergillosis among intubated patients with COVID-19: a prospective study. Clin Infect Dis. 2021;73(11):3606–3620.
- Strausz S, Kiiskinen T, Broberg M, FinnGen, et al. Sleep apnoea is a risk factor for severe COVID-19. BMJ Open Resp Res. 2021;8(1):e000845.
- Nori P, Cowman K, Chen V, et al. Bacterial and fungal coinfections in COVID-19 patients hospitalized during the New York city pandemic surge. Infect Control Hosp Epidemiol. 2021;42(1):84–88.
- Vaughn VM, Gandhi TN, Petty LA, et al. Empiric antibacterial therapy and community-onset bacterial coinfection in patients hospitalized with coronavirus disease 2019 (COVID-19): a multi-hospital cohort study. Clin Infect Dis. 2021;72(10):e533–e541.

