Abstract
Background
Pyogenic liver abscess (PLA) is a rare but potentially life-threatening disease, and estimates suggest a gradual increase in the incidence during the last decades. The primary aim of this study was to report the incidence, trend and aetiology of PLA during a decade in Southern Sweden.
Methods
This was a population-based observational cohort study between 2011 and 2020 in Skåne, Southern Sweden. Data were retrieved from the Swedish National Board of Health and Welfare for all individuals diagnosed with liver abscess (K750) according to ICD-10 (International Statistical Classification of Diseases, 10th revision).
Results
A total of 456 episodes of PLA occurred in 364 patients during the study period. The median age of the first PLA episode was 71 years (range 3–97) and 57% (n = 206) were men. The mean incidence of all patients was 3.4/100,000 person-years (range 1.8–5.2). The incidence increased almost three times, from 1.8/100,000 person-years in 2011 to 5.2/100,000 person-years in 2020. Streptococcus species, Escherichia coli and Klebsiella species accounted for the vast majority of both mono- and polymicrobial findings in both blood and local abscess cultures. 16s rDNA added information about aetiology in 37% of episodes.
Conclusion
The incidence of PLA increased during the study period, and Streptococcus spp., Klebsiella spp. and E. coli dominated both blood and local cultures. Despite antimicrobial therapy, pathogens could be found in local abscess cultures several weeks into treatment. Increased use of 16s rDNA in the management of PLA could be beneficial.
Introduction
Pyogenic liver abscess (PLA) is a rare but potentially life-threatening disease, and there have previously only been a few population-based studies on the incidence and aetiology of liver abscesses. Estimates suggest a gradual increase in the incidence during the last decades, which could possibly be due to improvements in diagnosis as well as an ageing population [Citation1–4].
Although studies presenting population-based incidences of PLA are lacking from parts of the world, the incidence seems to differ geographically. In Denmark, Canada and the United States, reported incidences are 1.0, 2.3 and 4.1 per 100,000 person-years, respectively [Citation1,Citation3,Citation5]. Higher estimates have been reported from Asia, with incidences of 14.4 and 17.6 per 100,000 person-years in South Korea and Taiwan, respectively [Citation2,Citation6].
The aetiology of PLA varies by geography. In North America, Escherichia coli is the most frequently found organism, whereas in addition to E. coli, Staphylococcus species and Streptococcus species are the dominating pathogens in Europe [Citation7,Citation8]. In contrast, multiple studies from Asia report that Klebsiella spp. are the dominating pathogens [Citation9–12]. Fusobacteria and Bacteroides are anaerobic bacteria associated with PLA, although anaerobic aetiology is rare and mainly associated with opportunistic infections [Citation13]. However, this could be explained by difficulties in culturing anaerobes, which could lead to underreporting [Citation14,Citation15]. Anaerobic bacteria can be detected by analysis of the gene coding for 16S ribosomal DNA (16S rDNA) using polymerase chain reaction (PCR). Also, 16S rDNA can be used after antimicrobial treatment has been initiated, since DNA from dead bacteria is detected. Previously, PCR has primarily been used to determine virulence factors of Klebsiella spp. in PLA, and detailed accounts of detecting causative bacteria using 16S rDNA are scarce [Citation16].
This study is adding to the few, population-based studies on incidence and aetiology of PLA and provides an updated perspective. In addition, detailed reports of PLA, taking into account both blood and local abscess cultures in relation to the timing of initiated antimicrobial treatment, have not previously been published.
The primary aim of this study was to investigate the incidence, trend and aetiology of PLA during a decade in Southern Sweden. A secondary aim was to investigate if 16S rDNA combined with standard cultures, could give additional information on the aetiology of PLA.
Methods
Study design and setting
This population-based observational cohort study included all individuals in Skåne, a region in Southern Sweden with a population of 1.4 million, diagnosed with liver abscess, between January 1, 2011 and December 31, 2020. Medical records of included individuals were audited according to a predefined study protocol, including age, sex, symptoms, immunosuppression, vital signs, comorbidities, smoking habits, laboratory test results, radiological imaging and microbiological analysis.
Data sources
Data (personal identification numbers) were retrieved from the Swedish National Board of Health and Welfare for all individuals diagnosed with liver abscess (K750) according to ICD-10 (International Statistical Classification of Diseases, 10th revision) between 2011 and 2020. The medical records of all patients were reviewed using the software system Melior (Melior, Siemens Healthcare Service, Upplands Väsby, Sweden). The database at the Department of Clinical Microbiology in Lund is responsible for all microbiological diagnostics in the region covering both public and private clinics, including hospitals, outpatient clinics and primary care. Laboratory Registers were accessed through the Laboratory Information Management System ww-lab (Autonik, Nyköping, Sweden).
Participants
Patients of all ages were included, and both in-hospital and outpatient care were included. The diagnosis K750 could be either main or secondary diagnosis. Patients misdiagnosed with PLA (i.e. the abscess was located outside the liver parenchyma), with liver metastases instead of PLA, with no documented abscess, with abscess caused by cestodes or amoebas were excluded. If medical records were lacking, or if patients resident outside Skåne were admitted in Skåne but directly transferred to a hospital outside Skåne, patients were excluded. A PLA episode was defined as a patient diagnosed with and treated for a PLA, not including relapses or recurrences within 365 days. Thus, only one incident PLA case per patient per year was included, and incidence and corresponding microbiological aetiology for this case were presented.
Data variables
All medical records were reviewed according to a predefined study protocol, including age, sex, main symptom and duration of symptoms, from where the patient was admitted, immunosuppression and vital signs. Vital signs were used to calculate National Early Warning Score (NEWS2, http://lof.se/filer/NEWS2-broschyr.pdf) and quick Septic related Organ Failure Assessment (qSOFA) [Citation17]. Comorbidities were assessed using Charlson’s comorbidity index [Citation18]. Current smoking was registered, as were C-reactive protein (CRP), lactate, white blood cell (WBC) count, procalcitonin, erythrocyte sedimentation rate (ESR), bilirubin, alanine aminotransferase (ALT), alkaline phosphatase (ALP), albumin and prothrombin time/international normalised ratio (PT/INR). Laboratory tests were collected within 24 h of the radiological discovery of PLA. The radiological entity, magnetic resonance imaging (MRI), computed tomography (CT) or ultrasound used to diagnose PLA was reported and refers to the entity used when PLA was first suspected. Data on baseline characteristics were assembled from the patients’ first admission due to PLA.
Microbiological analyses
For blood cultures, the BacT/ALERT® (bioMérieux, Inc., Marcy-l'Etoile, France) blood culture system was used until December 2014, when it was replaced by the BACTEC™ FX (Becton Dickinson, Franklin Lakes, NJ, USA), both using a 5-day incubation standard and both aerobic and anaerobic bottles.
For biopsies and aspirated material, the standard culture method during the study period consisted of inoculation of part of the sample into tryptic soy broth incubated in CO2-enriched atmosphere, as well as direct plating of the sample on blood agar incubated in CO2-enriched atmosphere and fastidious anaerobe agar incubated under anaerobic conditions. Additional agar plates and broths were added in selected cases depending on the diagnostic questions asked and at the discretion of the clinical microbiologists responsible for the analysis. The main method for bacterial and fungal species identification during the study period was matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS, Bruker Daltonics), supplemented by 16s and ITS sequencing using Sanger methodology as the main second line method when species identification using MALDI-TOF MS was unsuccessful.
Susceptibility testing was performed according to EUCAST methodology [Citation19], mainly by disc diffusion though supplemented by gradient tests.
Monomicrobial findings were one species of microbes obtained in cultures, whereas in polymicrobial findings two or more species were found. Blood and local abscess cultures represent the first culture obtained for each episode; additional cultures were not included in the study. A positive culture refers to a pathogen found, a negative culture thus denotes that no pathogen was identified.
Local abscess cultures were primarily obtained by aspiration or drainage of abscess, but for relapses acquired from an existing catheter.
On request, 16S rDNA and ITS PCR were done directly on the aspirated sample. Lysozyme, lysostaphin and proteinase K were used for DNA extraction. After amplification, Sanger sequencing was used for species identification.
Statistical analysis
Numerical data were presented with medians, means and range and qualitative data as proportions (%). Crude incidences of PLA in Skåne were calculated by dividing the number of annual episodes by the number of inhabitants in Skåne for each year, obtained from the open database of Statistics Sweden [Citation20]. The population at the end of June each year was used as the population for that particular year. Annual incidence rates were age- and sex-standardized to the population of south Sweden in 2020 (the most recent population in south Sweden) to adjust for changes in population demographics over time, including 95% confidence intervals. All statistical analyses were performed using Prism version 7 (GraphPad).
Ethics
This study was granted ethical approval from the Ethical Review Board (DNR-2020-06526). Informed consent was waived due to the observational and retrospective study design.
Results
Characteristics of included patients
In total, 364 patients were included in the study (). The median age of included patients was 71 years (range 3–97) and 57% (n = 206) were men. The median Charlson comorbidity score was 5 (range 0–15) and 23% (n = 84) of patients were immunosuppressed. A detailed list of baseline characteristics is found in .
Figure 1. Flowchart of steps of inclusion. aICD diagnosis K750. bPatients were diagnosed with K750, but no record of abscess could be found. cNon-Skåne residents, transferred to hospitals outside of Skåne. dThe diagnosis K750 was not present in the medical record. PLA: pyogenic liver abscess.
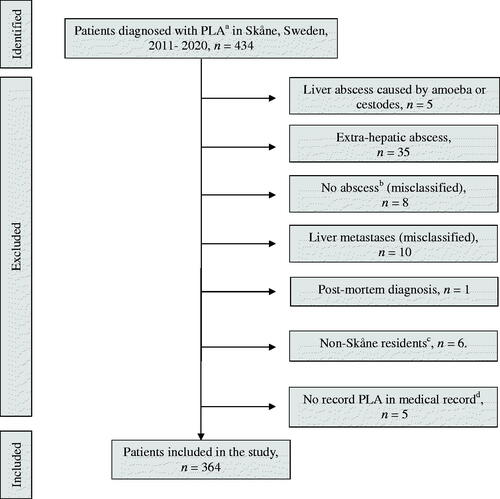
Table 1. Baseline characteristics and information gathered from the first admission.
Incidence of PLA during the study period
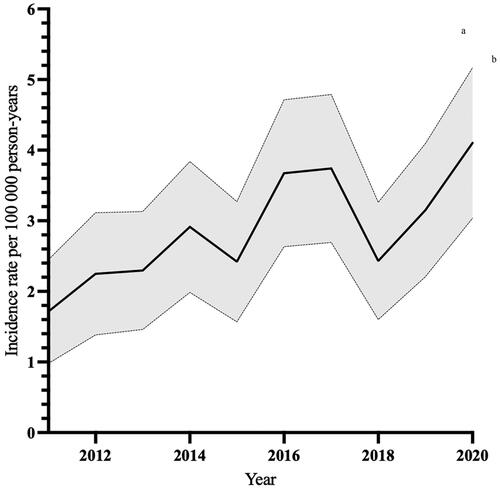
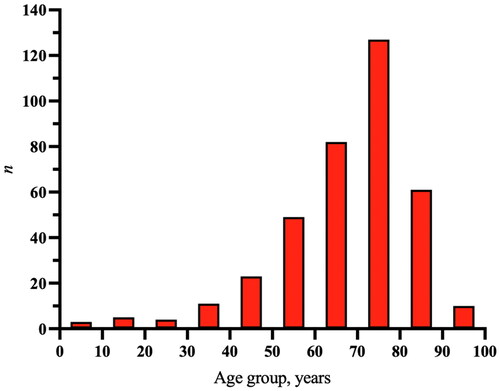
There were 375 incident cases of PLA in 364 patients during the study period. The mean age- and sex-standardized PLA incidence was 2.9/100,000 person-years (95% CI 2.0–3.8) between 2011 and 2020. The incidence more than doubled over the study period, from 1.7/100,000 person-years (95% CI 1.0–2.5) in 2011 to 4.1/100,000 person-years (95% CI 3.0–5.2) in 2020 ( and Supplementary Table A1). The age groups 61–70 and 71–80 years accounted for the majority of incident episodes (n = 209, 56%; ).
Symptoms and diagnostic method
The main symptoms were fever (n = 166, 44%), abdominal pain (n = 115, 31%) and fatigue (n = 25, 7%). Out of all patients, 58% (n = 217) reported abdominal pain at admission (). The majority (83%, n = 311) were diagnosed by CT, followed by ultrasound (15%, n = 56) and MRI (2%, n = 8).
Table 2. Clinical findings of all included PLA episodes (n = 375).
Based on imaging, 61% (n = 230) presented with a solitary abscess, 38% (n = 144) with multiple abscesses, and for one episode this information was unavailable. The median abscess diameter was 6 cm (range 0.8–25) and in 68% (n = 254) of episodes, the liver abscess was located in the right lobe and in 18% (n = 69) in the left lobe. Both lobes were involved in 11% (n = 42) of episodes and in 3% (n = 10) localisation was not documented. A portal thrombosis was concurrently found in 11% (n = 42) of PLA episodes ().
Table 3. Laboratory characteristics and diagnostic modalities of all PLA admissions.
Microbiology – blood cultures
Blood cultures were obtained in 93% (n = 350) of all episodes of PLA and 54% (n = 188) were positive (Supplementary Table A2). Monomicrobial results were found in 73% (n = 138) of blood cultures, and the most common pathogens were Streptococcus spp. 29%, n = 40), E. coli (19%, n = 26) and Klebsiella spp. (17%, n = 24). The most common Streptococcus species belonged to the Streptococcus anginosus complex (n = 37, 93%). Klebsiella species found were Klebsiella pneumoniae (15%, n = 21), Klebsiella oxytoca group (1%, n = 2) and Klebsiella aerogenes (1%, n = 1; Supplementary Table A3).
Polymicrobial findings accounted for 26% (n = 49) of positive blood cultures. In most cases, both Gram-negative and -positive bacteria were isolated (71%, n = 35). In 20% (n = 10) only Gram-negative bacteria were found and in 8% (n = 4) only Gram-positive. Candida spp. were found in 4% (n = 2) of all episodes. The most frequent pathogens in polymicrobial infections were E. coli (53%, n = 26), Klebsiella spp. (47%, n = 23), Streptococcus spp. (35%, n = 17), Enterococcus spp. (29%, n = 14), Clostridium spp. (22%, n = 11) and Bacteroides spp. (12%, n = 6; Supplementary Table A4).
Of isolated E. coli and Klebsiella species in blood cultures, 6% (n = 3) and 4% (n = 2) were ESBL-producing, respectively.
Microbiology – abscess cultures
Abscess cultures were obtained from 61% (n = 228) of episodes, and in 75% (n = 172), at least one pathogen was isolated. The most frequent monomicrobial findings were Streptococcus spp. (32%, n = 32), E. coli (21%, n = 21) and Klebsiella spp. (13%, n = 13; Supplementary Table A3). The most common Streptococcus species was the S. anginosus complex (n = 30, 94%). Klebsiella species were K. pneumoniae (n = 12, 12%) and K. oxytoca group (n = 1, 1%).
Abscess cultures yielded 41% (n = 71) polymicrobial results. In 63% (n = 45) both Gram-negative and Gram-positive bacteria were isolated. Only Gram-positive bacteria were found in 1% (n = 1), only Gram-negative in 17% (n = 12) and Candida spp. in 17% (n = 12; Supplementary Table A4). In abscess cultures, the most common microbes were Enterococcus spp. (42%, n = 30), E. coli (45%, n = 32), Klebsiella spp. (34%, n = 24), Streptococcus spp. (27%, n = 22) and mixed anaerobic flora (21%, n = 15; Supplementary Table A4). In abscess cultures, ESBL production was detected in two E. coli strains (4%) and one K. pneumoniae (3%; Supplementary Table A3).
Microbiology – 16S rDNA
Of all 375 PLA episodes, 16S rDNA was analysed in 26% (n = 98) of abscess samples. Compared with abscess cultures, 16S rDNA gave additional information in 37% (n = 36) of episodes. In 50% (n = 49), 16S rDNA results were identical to those of abscess culture and in 13% (n = 13) 16S rDNA was negative.
Antimicrobial therapy
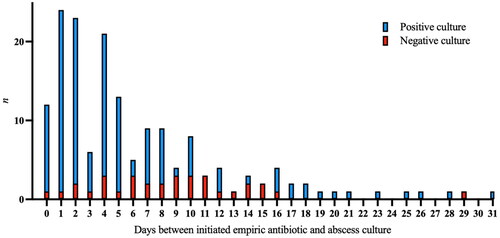
The median time from admission to administration of empiric antibiotics was 0 days (range 0–17, n = 372). In three episodes, antimicrobials were never initiated. After antimicrobial therapy was initiated, the number of positive abscess cultures decreased over time but cultures could still be positive weeks into antimicrobial treatment ().
Discussion
In this population-based observational study, we aimed to report the incidence, trend and aetiology of PLA during a decade in Skåne, Southern Sweden. Also, we wanted to investigate if 16S rDNA, combined with standard cultures, could give additional information on the aetiology of PLA.
Increasing incidence
We found that, although a rare infection, the age- and sex-standardized incidence of PLA increased during the study period. The mean incidence in our study (2.9/100,000 person-years) is in accordance with other population-based studies from North America, and slightly higher than in a study from Denmark [Citation1, Citation3, Citation5]. In Asia, crude incidence rates of PLA are much higher, attributable to endemic strains of Klebsiella [Citation9–11]. The PLA incidence more than doubled during the decade studied, and since the incidence was age and sex-standardized, this increase cannot be explained by an ageing population. We believe that the incidence-increase may be explained, at least in part, by increased diagnostic testing or access to radiology-guided aspiration. Also, as ascertainment of cases was based on diagnostic coding, improved coding during the study period could also affect the incidence.
The increased PLA incidence is not likely explained by an unhealthier population, as the median Charlson comorbidity index of patients for each year remained stable during the study period (Supplementary Table A5). Instead, the number of patients with a Charlson comorbidity index of zero increased during the study. This may suggest that the increased incidence is due to other factors, such as more virulent bacteria causing PLA in otherwise healthy patients, or that risk factors not included in the Charlson comorbidity index (such as surgery) could explain this development.
Aetiology of PLA
Our study shows that the aetiology of PLA is heterogenous, with a diversity of isolated anaerobic, aerobic, Gram-positive and Gram-negative and mono- and polymicrobial pathogens. However, Streptococcus spp. (dominated by the S. anginosus complex), E. coli and Klebsiella spp. (dominated by K. pneumoniae) accounted for the vast majority of both mono- and polymicrobial findings in both blood and abscess cultures. This is in line with previous reports [Citation3, Citation21]. Enterococcus species and anaerobic bacteria, such as Bacteroides- and Clostridium species, were also prevalent in both mono- and polymicrobial blood cultures. Consequently, it is important to cover these pathogens in empirical treatment. Thus, piperacillin/tazobactam or a third-generation cephalosporin, such as cefotaxime, combined with metronidazole are reasonable alternatives for empirical treatment of PLA. Enterococci and Candida species were more common in abscess cultures than in blood cultures, which could be explained by lengthy treatment with broad-spectrum antimicrobials which could select for these pathogens in local cultures. The prevalence of ESBL-producing Enterobacterales (EPE) was very low, in accordance with the low-endemic situation in Sweden regarding multidrug-resistant bacteria compared to other regions of the world [Citation22]. However, even in high-endemic settings, the prevalence of antibiotic resistance in Klebsiella spp. is low [Citation23].
Candida species were rarely isolated in our study, and, in our opinion, empirical coverage of EPE and Candida spp. is not needed.
The utility of 16S rDNA in aspirated abscess material has been previously demonstrated, especially in settings with a high prevalence of liver abscesses caused by Entamoeba histolytica [Citation24]. We believe that analysis of the 16S rDNA-gene from abscesses could be beneficial, as PCR added information in 37% of episodes in our study compared to standard culturing methods. PLAs are frequently caused by anaerobic bacteria, which are often difficult to culture [Citation15]. However, 16S rDNA does not allow susceptibility testing which is a disadvantage.
Aspiration and drainage of pus from PLA can be therapeutic, as primary source control [Citation25]. Our study clearly shows the utility of culture and 16S rDNA performed on abscess contents; although antimicrobials in many episodes had been administered; clinically relevant pathogens were found in abscess cultures several weeks into treatment (). This highlights the need for early abscess drainage, for therapeutic and diagnostic reasons and may justify the long antimicrobial treatment associated with this condition.
Strengths and limitations
To the best of our knowledge, this study includes one of the biggest cohorts of patients with PLAs, including detailed data on aetiology from both blood and abscess cultures. Other strengths of this study include the population-based study design. However, there are some potential weaknesses. First, the retrospective study design, with the risk of selection bias. Second, relying exclusively on ICD codes could underestimate the true incidence of PLA, as patients might be misdiagnosed. Third, some abscess cultures were obtained after initiation of antimicrobials and from catheters having been present >24 h, which could influence the microbiological results. Furthermore, the ascertainment of cases was based only on diagnostic coding. To validate that the incidence of PLA is indeed increasing, a second measure to screen for cases, such as cultures sent to the Department of Clinical Microbiology in Lund, could have been used. Unfortunately, we were unable to search the microbiological database for liver abscess samples only, as there is no specific code for liver abscess samples. Therefore, we could not investigate if a positive liver abscess culture was found with or without an associated diagnostic code of PLA (K750).
Although still rare, PLA is an emerging infection, and the incidence can be expected to increase further in the future with an ageing population. The results of this study may assist clinicians to diagnose and treat PLA and form a basis for empirical antimicrobial treatment of PLA in our region. More studies are needed to investigate why the PLA incidence is increasing and to evaluate optimal treatment.
Conclusion
The incidence of PLA increased during the study period, and Streptococcus spp., Klebsiella spp. and E. coli dominated both blood and local cultures. Despite antimicrobial therapy, pathogens could be found in local abscess cultures several weeks into treatment. Increased use of 16s rDNA in the management of PLA could be beneficial.
Author contributions
O.L. conceived the study, performed data acquisition and administered the project. The study design methodology was finalised by A.B., A.K. and O.L. Data curation, analysis and visualisation were performed by E.S., A.J., A.B., T.S. and O.L. The manuscript was initially drafted by O.L., E.S. and A.J. and critically revised by A.K., T.S. and A.B. All authors approved the final version of the manuscript.
Supplemental Material
Download MS Word (23.4 KB)Acknowledgements
The authors thank Dr. Gustav Torisson for valuable inputs on the statistical analyses.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Jepsen P, Vilstrup H, Schonheyder HC, et al. A nationwide study of the incidence and 30-day mortality rate of pyogenic liver abscess in Denmark, 1977–2002. Aliment Pharmacol Ther. 2005;21(10):1185–1188.
- Yoo JJ, Lee TK, Kyoung DS, et al. A population-based study of pyogenic liver abscess in Korea: incidence, mortality and temporal trends during 2007–2017. Liver Int. 2021;41(11):2747–2758.
- Kaplan GG, Gregson DB, Laupland KB. Population-based study of the epidemiology of and the risk factors for pyogenic liver abscess. Clin Gastroenterol Hepatol. 2004;2(11):1032–1038.
- Mohsen AH, Green ST, Read RC, et al. Liver abscess in adults: ten years experience in a UK Centre. QJM. 2002;95(12):797–802.
- Meddings L, Myers RP, Hubbard J, et al. A population-based study of pyogenic liver abscesses in the United States: incidence, mortality, and temporal trends. Am J Gastroenterol. 2010;105(1):117–124.
- Tsai FC, Huang YT, Chang LY, et al. Pyogenic liver abscess as endemic disease, Taiwan. Emerg Infect Dis. 2008;14(10):1592–1600.
- Cerwenka H. Pyogenic liver abscess: differences in etiology and treatment in Southeast Asia and Central Europe. World J Gastroenterol. 2010;16(20):2458–2462.
- Kurland JE, Brann OS. Pyogenic and amebic liver abscesses. Curr Gastroenterol Rep. 2004;6(4):273–279.
- Lok K-H, Li K-F, Li K-K, et al. Pyogenic liver abscess: clinical profile, microbiological characteristics, and management in a Hong Kong hospital. J Microbiol Immunol Infect. 2008;41(6):483–490.
- Wong W-M, Wong BCY, Hui CK, et al. Pyogenic liver abscess: retrospective analysis of 80 cases over a 10-year period. J Gastroenterol Hepatol. 2002;17(9):1001–1007.
- Wang J-H, Liu Y-C, Lee SS-J, et al. Primary liver abscess due to Klebsiella pneumoniae in Taiwan. Clin Infect Dis. 1998;26(6):1434–1438.
- Lübbert C, Wiegand J, Karlas T. Therapy of liver abscesses. Viszeralmedizin. 2014;30(5):334–341.
- Jayan GS, Rajkumari N, Biswas R, et al. Detection of Entamoeba histolytica and bacterial etiological agents in patients with clinically suspected cases of liver abscesses. J Parasit Dis. 2022;46(1):254–261.
- Roediger R, Lisker-Melman M. Pyogenic and amebic infections of the liver. Gastroenterol Clin North Am. 2020;49(2):361–377.
- Ogah K, Sethi K, Karthik V. Clostridium clostridioforme liver abscess complicated by portal vein thrombosis in childhood. J Med Microbiol. 2012;61(Pt 2):297–299.
- Zhang S, Zhang X, Wu Q, et al. Clinical, microbiological, and molecular epidemiological characteristics of. Antimicrob Resist Infect Control. 2019;8:166.
- Singer M, Deutschman CS, Seymour CW, et al. The third international consensus definitions for sepsis and septic shock (sepsis-3). JAMA. 2016;315(8):801–810.
- Charlson ME, Pompei P, Ales KL, et al. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis. 1987;40(5):373–383.
- EUCAST. Clinical breakpoints – breakpoints and guidance. Version 13.0 [Internet]. [cited 2023 Feb 21]. https://www.eucast.org/clinical_breakpoints
- Folkmängden per månad efter region, ålder och kön. År 2000M01 – 2022M09 [Internet]. Statistiska centralbyrån [updated 2022 Nov 10; cited 2022 Dec 6]. https://www.statistikdatabasen.scb.se/pxweb/sv/ssd/START__BE__BE0101__BE010
- Tian LT, Yao K, Zhang XY, et al. Liver abscesses in adult patients with and without diabetes mellitus: an analysis of the clinical characteristics, features of the causative pathogens, outcomes and predictors of fatality: a report based on a large population, retrospective study in China. Clin Microbiol Infect. 2012;18(9):E314–E330.
- Aspevall O, Bergfeldt V, Billström H, et al. 2020. Sales of antibiotics and occurrence of antibiotic resistance in Sweden. Public Health Agency of Sweden and National Veterinary Institute; 2020. https://www.sva.se/media/8d9678c390929e9/swedres_svarm_2020.pdf
- Jun JB. Liver abscess. Infect Chemother. 2018;50(3):210–218.
- Reyna-Fabián ME, Zermeño V, Ximénez C, et al. Analysis of the bacterial diversity in liver abscess: differences between pyogenic and amebic abscesses. Am J Trop Med Hyg. 2016;94(1):147–155.
- Lardière-Deguelte S, Ragot E, Amroun K, et al. Hepatic abscess: diagnosis and management. J Visc Surg. 2015;152(4):231–243.