Abstract
Background
The transition to PCR-based diagnosis of bacterial gastroenteritis (BGE) can increase the sensitivity but might reduce the clinical specificity. The aims of this study were (1) to compare the effect of the change from culture to PCR-based diagnostics on the reported incidence and positivity rates of BGE due to Salmonella, Shigella and Campylobacter species and (2) to compare the demographics, medical background, clinical characteristics and pre-analytic variables between cases with PCR-positive, culture-negative samples to cases with PCR-positive, culture-positive samples.
Methods
The study was performed at the Emek Medical Centre that serves a population of 0.5 million people in Northern Israel. The study included two parts: (1) a retrospective cohort study, comparing the incidence and positivity rates of laboratory-diagnosed BGE from January 2016 until December 22nd, 2019 when culture was the sole method to January 2020 until April 2023 when PCR was used; (2) a prospective cohort study, conducted between November 2020 until April 2023 that compared the demographics and clinical characteristics of BGE cases that were diagnosed by PCR alone versus cases that were diagnosed by both PCR and culture.
Results
The incidence rate between-periods comparability ratio was only 113% since the incidence rate did not increase during 2020, the first year of the COVID-19 pandemic. The sample positivity rate increased since 2020, with between-periods comparability ratio of 159%. In the second period, the sample positivity rates of culture vs. PCR alone differed between the pathogens and were 90.2%, 63.8% and 54.2% for Salmonella, Campylobacter and Shigella species, respectively (p < 0.001). The following variables were identified as independent predictors of culture positivity: (1) Salmonella infection (O.R. = 10.6, 95% C.I. 3.6–31.1, p < 0.001); (2) Shigella infection (O.R. = 0.46, 95% C.I.0.23–0.93, p = 0.032); (3) time from sample submission to culture (O.R.=0.73, 95% C.I. 0.58–0.92, p = 0.008); (4) the presence of abdominal pain (O.R. = 1.98, 95% C.I. 1.04–3.79, p = 0.038) and the PCR mean Ct value (O.R. = 0.89, 95% C.I.0.85–0.94, p < 0.001).
Conclusions
The use of PCR had led to improved sensitivity, without noticeable decrease in the clinical specificity. This was especially important in the case of the more fastidious organisms.
Keywords:
Introduction
Diarrheal diseases is a major hurdle to public health worldwide, especially in developing countries [Citation1]. Non-Typhoidal Salmonella (NTS), Shigella or Campylobacter species are identified as the most common bacterial aetiologies for this disease, henceforth bacterial gastroenteritis (BGE) [Citation1]. In Israel, prior to the COVID-19 epidemic, the incidence rate of NTS had initially declined, followed by a transient increase in 2017 [Citation2], the incidence rate of Campylobacter had increased until 2009 and remained stable ever since while the incidence rate of Shigella, the least common of these agents had remained stable [Citation3].
The laboratory identification of BGE was traditionally based on stool culture with specific media and incubation conditions for NTS, Shigella or Campylobacter [Citation4]. In recent years, PCR-based methods become widely available in clinical laboratories. These methods, although typically more sensitive than culture [Citation5], have the potential drawback of clinical false-positive, i.e. the inability to distinguish between the presence of a viable pathogen and either asymptomatic carriage or merely DNA remnants [Citation6]. If true, this phenomena might lead to unnecessary medical measures, such as antimicrobial therapy and isolation of patients. The Emek Medical Centre (EMC) laboratory had shifted in December 2019 from culture to PCR-based diagnostics of BGE. Therefore, it was of paramount importance to assess the effects of this change on the sample’s positivity rates of the different BGE pathogens and the reported incidence rates and to examine the validity of the concern for clinical false-positive reports due to this change.
The aims of this study were (1) to compare the effect of the change from culture-based to PCR-based diagnostics on the reported incidence and positivity rates of BGE due to Salmonella, Shigella and Campylobacter species and (2) to compare the demographics, medical background, clinical characteristics and sample’s pre-analytic variables between cases with PCR-positive, culture-negative samples to cases with PCR-positive, culture-positive samples. Our hypotheses were (1) that the reported incidence and positivity rates of BGE will increase in the second period and (2) that the cases with PCR-positive, culture-negative samples will have milder clinical course compared with the PCR-positive, culture-positive cases.
Methods
Setup and population
The study was performed at the Microbiology Laboratory and the Infectious Diseases Unit of the Emek Medical Centre (EMC) in Afula, Israel. The EMC is part of the Clalit Health Service (CHS) and the laboratory serves as the regional laboratory of its members. The region of North-East Israel includes a population of about 0.5 million of Clalit health members, made up of equal numbers of Arabs and Jews living predominantly in rural settlements
Since the last week of December 2019, the EMC laboratory replaced the routine use of culture for the diagnosis of bacterial gastroenteritis to PCR-based diagnosis, using cultures as add-on test for samples that were positive by PCR to Shigella, Salmonella, or Campylobacter (see methods below). The study included all outpatient cases of laboratory-diagnosed bacterial gastroenteritis (Shigella, Salmonella, or Campylobacter) of any age.
Study design
The study included two parts: (1) a retrospective cohort study, comparing the incidence and the positivity rates of laboratory-diagnosed bacterial gastroenteritis from January 2016 until December 22nd, 2019 when culture was the sole method to January 2020 until April 2023 when PCR was used (the last week of December 2019 was regarded as a washout period); (2) a prospective cohort study, conducted between November 2020 until April 2023 that compared the demographics and clinical characteristics of bacterial gastroenteritis cases (due to Shigella, Salmonella, and campylobacter species) that where diagnosed by PCR alone versus cases that were diagnosed by both PCR and confirmatory culture. Additionally, we compared pre-analytic factors pertaining to sample processing between the two groups.
Eligible cases were first approached by phone by an Infectious Disease physician (authors HH and CK) and were asked for their consent to participate in the study. Consented patients were then approached by the study first author (DS) and underwent a phone questionnaire (see supplemental material) that included sociodemographic data, general medical background, diarrheal disease characteristics and other symptoms, food and animal exposure, antibiotic exposure, and the timing of sample collection in relation to onset of diarrhoea. The study was approved by the ethical review board of the EMC.
Laboratory methods
Stool samples were transported from the community clinics and were tested daily except on weekends, where they were refrigerated at 4 °C until tested. Stool samples were suspended in ASL buffer (Qiagen, Hilden, Germany) followed by DNA extraction that was conducted using the STARMag Universal Cartridge Kit (Seegene, Duesseldorf, Germany) on the STARLET automated extraction platform (Seegene). Bacterial enteropathogens were tested by the Allplex™ GI -Bacteria(I) PCR assay (Seegene, Seoul, South Korea) [Citation7].
The assay detects seven enteropathogens including Shigella spp./Enteroinvasive Escherichia coli (EIEC), Salmonella spp., Campylobacter spp., Aeromonas spp., Yersinia enterocolitica, Vibrio spp. and Clostridium difficile toxin B. The PCR Ct values were documented in positive samples, in order to assess the quantity of the organisms in the sample and its relation with culture positivity. Samples that were positive by PCR for Shigella, Salmonella, or Campylobacter were cultured within the same 24 h of the PCR results. Shigella/EIEC and Salmonella PCR-positive samples were cultured on SS agar plates and Selenite broth (Hylabs, Rehovot, Israel) and incubated at 36 °C for 24 h. Campylobacter PCR-positive samples were cultured on Campylobacter Medium blood-free charcoal agar plates (Hylabs, Rehovot, Israel) and incubated in microaerophilic conditions at 42 °C for 48 h. Suspicious colonies for Shigella and Salmonella were identified using Kligler’s Iron Agar ENTEROPLUS and INDOL CAPS (NOVAmed, Jerusalem, Israel) test and subtyped by agglutination sera test (Remel Europe Ltd). Isolation of EIEC was not attempted as this was never the practice in our lab or in Israel in general. Suspicious colonies for Campylobacter were identified by MALDI Biotyper Sirius system (Bruker Daltonics, Bremen, Germany).
Statistical analysis
Trend analysis was done using the Joinpoint Regression Program version 4.7.0.0 [Citation8,Citation9] that included the following analyses: (1) basic trend analysis expressed as average percent change (APC) and joinpoint identification (i.e. the identification of points of trend change); (2) trend analysis with ‘‘jump model’’ set at 2020 (i.e. the replacement of the testing method from culture to PCR), to determine whether the overall trend was affected by the methodological change.
The two study groups were compared using the Chi2 test for categorical variables and the t-test for continuous variables. Multivariate analysis for the identification of independent predictors of culture positivity was done using logistic regression and included all variables identified with a p-value < 0.05 in the univariate analysis.
Results
Laboratory-reported bacterial gastroenteritis incidence and sample positivity rates
The trends in Laboratory-reported bacterial gastroenteritis incidence and sample positivity rates between 2016 to April 2023 are presented in and . Campylobacter species were the most common pathogen in both the pre-PCR period (2016–2019) and the PCR period (2020–2023). Of the 1376 Campylobacter that were isolated by culture between 2020–2023, 1173 (85%) were C. jejuni and 161 (12%) were C. coli. Shigella infection was more common than Salmonella in the pre-PCR period (2016–2019, with the exception of 2017). Between 2020–2022, which corresponded with the COVID-19 pandemics period, Salmonella had become more common; in 2023 Shigella infection were again more common than Salmonella.
Figure 1. Trends in laboratory-reported bacterial gastroenteritis incidence and sample positivity rates, 2016–April 2023. The transition (‘jump’) point from culture to PCR-based testing was at 2020. (A) Annual incidence rates. (B) Annual sample positivity rates. P-values were >0.05 for the models.
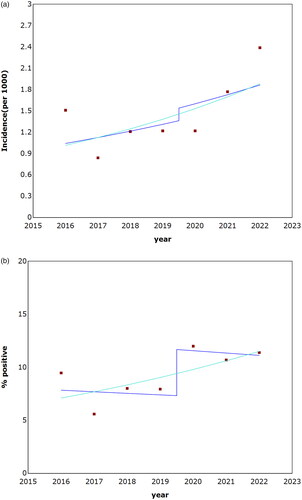
Table 1. Trends of laboratory-reported bacterial gastroenteritis incidence rates and sample positivity rates, January 2016–April 2023.
The annual incidence fluctuated in both the pre-PCR and the PCR period (); most notably in 2020, being much lower compared with 2021–2022. Assuming a 'jump’ point in 2020, the model estimated the comparability ratio (of post/pre change) as 113% with an annual percentage change (APC) of 7.9%, without a jointpoint.
The annual sample positivity rate () fluctuated in the pre-PCR period, similar to the changes in the annual incidence (); the rates were stable in the PCR period. Assuming a 'jump’ point in 2020, the model estimated the comparability ratio (of post/pre change) as 159% without a jointpoint but unlike the incidence rate trend, the APC was declining at a rate of −1.92%.
Pre-analytical and microbiological characteristics of PCR-positive bacterial gastroenteritis cases
In the PCR period (2020–2023), primary testing was done by PCR with subsequent culturing of PCR-positive samples. The pre-analytical and microbiological characteristics of PCR-positive bacterial gastroenteritis cases are presented in . The positivity rates of culture vs. PCR only differed between the pathogens and was 90.2%, 63.8% and 54.2% for Salmonella, Campylobacter and Shigella species, respectively (p < 0.001).
Table 2. Pre-analytical and microbiological characteristics of PCR-positive bacterial gastroenteritis cases.
The two time-related variables, time from symptom onset to sample submission and from that point to culture inoculation showed significant associations between shorter duration and higher positivity rate. Higher PCR Ct values were significantly associated with lower positivity rate, albeit the difference in the mean Ct value was only 1.81 cycles.
Demographic and clinical characteristics of PCR-positive bacterial gastroenteritis cases
Patients from both groups had similar demographic characteristics, with an average age of less than 21 years old and a low rate of underlying diseases (). The clinical features of the BGE itself were also similar with two exceptions: (1) patients in the PCR-positive, culture-negative group had longer total duration of illness and (2) patients in the PCR-positive, culture-positive group had a higher rate of abdominal pain. The majority of patients in both groups were treated with antimicrobials.
Table 3. Demographic and clinical characteristics of PCR-positive bacterial gastroenteritis cases.
Recent restaurant dining was common in both groups and there were no other differences in nutritional habits between the groups (Table S1).
We performed a multivariate analysis to identify the predicting variables for stool culture positivity that included the following variables: (1) the bacterial pathogen (with Campylobacter species as reference); (2) time from symptom onset to sample submission; (3) time from sample submission to culture inoculation; (4) total duration of illness; (5) abdominal pain and (6) PCR Ct values. The following variables were identified as independent predictors of culture positivity: (1) Salmonella infection (O.R.=10.6, 95% C.I. 3.6–31.1, p < 0.001); (2) Shigella infection (O.R.=0.46, 95% C.I.0.23–0.93, p = 0.032); (3) time (days) from sample submission to culture (O.R.=0.73, 95% C.I.0.58–0.92, p = 0.008); (4) the presence of abdominal pain (O.R.=1.98, 95% C.I.1.04–3.79, p = 0.038) and the PCR mean Ct value (O.R.=0.89, 95% C.I.0.85–0.94, p < 0.001). When the analyses were done separately for each pathogen, both the time from sample submission to culture and the PCR mean Ct values were found to be a statistically significant predictors for culture positivity for Campylobacter and Shigella infections (data not shown). In Salmonella infections, where the rate of culture positivity was the highest (90.2%, ), no variable was found as predictive of culture positivity.
Discussion
Methodological changes can lead to significant alterations in the reported incidence rates of infectious diseases. Microbiologists and physicians alike are typically aspiring to improve the analytical sensitivity of microbiological assays and thus the transformation to PCR offers an attractive alternative to culture-based or even antigen-based diagnostics. However, such transition is not without its risk, as the increase in the analytical sensitivity might lead to a lower ‘clinical’ specificity and thus to a lower positive predictive value. Examples for these conditions can be found in both gastrointestinal infections (e.g. C. difficile [Citation10]) and in other types of infections (e.g. Pneumocystis jirovecii [Citation11]). With that in mind, our hypotheses were that the transition to PCR-based detection of BGE will lead to (1) an increase in the reported incidence of BGE that will be combined with (2) changes in the clinical characteristics of PCR-positive cases, with milder or even asymptomatic cases being diagnosed. Surprisingly, both of these hypotheses were not found to be accurate.
Previous studies have found an increase in BGE reports following the transition to PCR-based detection [Citation12,Citation13]. Our findings were more equivocal and varied in different years and with different pathogens. As the first year of the transition, 2020, was also the first year of the COVID-19 pandemic, the incidence rate of PCR-detected BGE had not increased compared with the pre-transition period whereas the culture-based incidence had declined (), suggesting an actual decline in the incidence. The latter phenomena was reported also on a national level in Israel [Citation14] as well as from other countries [Citation15,Citation16] and was most prominent in Shigella and thus is not likely to be due to methodological variations. The incidence rate of PCR-detected BGE had increased only in the second year of its implication (2021) with Shigella infection increasing only in 2022. As a result of this dynamics, the comparability ratio between the periods was only 118% () and thus the APC slope showed an increasing trend. Still, the sample positivity rate had increased already in 2020 with an overall comparability ratio was 157% resulting in a decline in the APC. Additional indication for the effect of this change was the rates PCR- positive, culture-positive samples (out of all PCR-positive samples) that was as low as 54.2% in Shigella infections (). Although this comparison was not perfect (since only positive samples were cultured), this low rate supports the hypothesis that the introduction of PCR had led to an increase in the reported incidence of BGE, especially of Shigella and Campylobacter species. While the PCR test for Shigella was capable of detecting EIEC as well (which was not isolated by our laboratory), it’s important to note that the occurrence of EIEC is generally lower in comparison to Shigella [Citation17]. Therefore, it is unlikely that EIEC would have had a significant impact on the overall incidence.
Our main concern following the introduction of PCR in 2020 was that it will lead to the identification of ‘clinical’ false positive cases, i.e. mildly symptomatic or even asymptomatic cases. This concern was based on the facts that gastrointestinal carriage may occur with Salmonella [Citation18], Campylobacter [Citation19] and even Shigella [Citation20]. Surprisingly, we have discovered that the clinical characteristics of patients with PCR-positive, culture-negative BGE were, for the most part indistinguishable from those with PCR-positive, culture-positive samples. The only clinical feature that differed between the groups was higher rate of abdominal pain in the culture-positive group. Other differences between the groups were mostly related to trivial microbiological differences, i.e. (1) higher rates of the more fastidious Shigella and Campylobacter compared with Salmonella in the PCR-positive, culture-negative group compared with Salmonella and (2) higher PCR Ct values in the PCR-positive, culture-negative group vs. the culture-positive group, as previously described [Citation21]. An intriguing difference between the groups was a longer duration of symptomatic illness prior to sample submission (in univariate analysis only) and longer duration from sample submission to culture. Similar finding was also reported in another study [Citation22] and can be explained by declining viability of the bacterial pathogens when sample submission and processing are delayed. Together, these findings suggests that at least in the case of Salmonella, Campylobacter and Shigella, the use of PCR had led to improved sensitivity, without noticeable decrease in the clinical specificity. This was especially important in the case of the more fastidious organisms and in cases where sample submission was delayed. Of note, culture might still have a role in determining antimicrobial susceptibility and for epidemiological surveillance.
The two parts of our study have innate limitations. First, since culture was done only in PCR-positive cases during the second period, it is possible that PCR-negative, culture-positive cases were missed and thus the actual incidence might have been higher. Second, as discussed above, determining the effect of methodological change on the reported incidence is a difficult task to begin with [Citation23] and was further hampered by the concurrent COVID-19 pandemic. Nevertheless, the effect of the change was apparent in both the change in the sample positivity rates and in comparison to the parallel confirmatory culture positivity rate during the second period (2000–2023). Third, the evaluation of ‘clinical’ specificity was always problematic due to the lack of definite clinical gold-standard. Still, the similarity in clinical features observed between the PCR-positive, culture-positive and the PCR-positive, culture-negative groups suggests that PCR can be used for the detection of BGE without undermining clinical specificity. Of note, the conclusions of our study might not be valid in areas with high rate of carriage of these organisms. Future studies may be necessary to evaluate the impact of the diagnostic transition on the utilisation of antimicrobials in BGE.
Supplemental Material
Download MS Word (17.3 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Troeger C, Blacker BF, Khalil IA, et al. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of diarrhoea in 195 countries: a systematic analysis for the global burden of disease study 2016. Lancet Infect Dis. 2018;18(11):1211–1228. doi: 10.1016/S1473-3099(18)30362-1.
- Bassal R, Davidovich-Cohen M, Yakunin E, et al. Trends in the epidemiology of Non-Typhoidal salmonellosis in Israel between 2010 and 2021. Int J Environ Res Public Health. 2023;20(9):5626. doi: 10.3390/ijerph20095626.
- Bassal R, Keinan-Boker L, Cohen D. A significant decrease in the incidence of shigellosis in Israel during covid-19 pandemic. Int J Environ Res Public Health. 2021;18:1–7.
- Leber AL. Clinical microbiology procedures handbook. 4th ed. Leber AL, editor. Washington, DC: ASM Press; 2016.
- Teh R, Tee WD, Tan E, et al. Review of the role of gastrointestinal multiplex polymerase chain reaction in the management of diarrheal illness. J Gastroenterol Hepatol. 2021;36(12):3286–3297. doi: 10.1111/jgh.15581.
- Binnicker MJ. Multiplex molecular panels for diagnosis of gastrointestinal infection: performance, result interpretation, and cost-effectiveness. J Clin Microbiol. 2015;53(12):3723–3728. doi: 10.1128/JCM.02103-15.
- Zimmermann S, Horner S, Altwegg M, et al. Workflow optimization for syndromic diarrhea diagnosis using the molecular seegene AllplexTM GI-Bacteria(I) assay. Eur J Clin Microbiol Infect Dis. 2020;39(7):1245–1250. doi: 10.1007/s10096-020-03837-4.
- Kim HJ, Fay MP, Feuer EJ, et al. Permutation tests for joinpoint regression with applications to cancer rates. Statist Med. 2000;19(3):335–351. doi: 10.1002/(SICI)1097-0258(20000215)19:3<335::AID-SIM336>3.0.CO;2-Z.
- Yavor A, Ben-Zvi H, Freeman S, et al. Institutional burden of Carbapenemase-Producing enterobacterales: the effect of changes in surveillance culture methodology. Microb Drug Resist. 2020;26(11):1350–1356. doi: 10.1089/mdr.2019.0478.
- van Dorp S, Hensgens MPM, Virolainen A; E.J. Kuijper on behalf of the ECDIS-Net Participants S., et al. Diagnostic testing and measurement of Clostridium difficile infections across Europe. Berlin: ECCMID; 2013. p. 1869.
- Sokulska M, Kicia M, Wesołowska M, et al. Pneumocystis jirovecii—from a commensal to pathogen: clinical and diagnostic review. Parasitol Res. 2015;114(10):3577–3585. doi: 10.1007/s00436-015-4678-6.
- States U, Iwamoto M, Huang JY, et al. Bacterial enteric infections detected by culture-independent diagnostic tests. Morb Mortal Wkly Rep. 2015;64:252–257.
- May FJ, Stafford RJ, Carroll H, et al. The effects of culture independent diagnostic testing on the diagnosis and reporting of enteric bacterial pathogens in Queensland, 2010 to 2014. Commun Dis Intell Q Rep. 2017;41(3):E223–E230.
- Israeli Center for Disease Control. Gastrointestinal Infections Weekly Report, April 2023. 2023. Available from: https://www.gov.il/BlobFolder/reports/maiim-29042023/he/files_weekly-intestine_maiim-29042023.pdf
- Ray LC, Collins JP, Griffin PM, et al. Decreased incidence of infections caused by pathogens transmitted commonly through food during the COVID-19 pandemic—foodborne diseases active surveillance network, 10 U.S. Sites, 2017–2020. MMWR Morb Mortal Wkly Rep. 2021;70(38):1332–1336. doi: 10.15585/mmwr.mm7038a4.
- Love NK, Elliot AJ, Chalmers RM, et al. Impact of the COVID-19 pandemic on gastrointestinal infection trends in England, February-July 2020. BMJ Open. 2022;12(3):e050469. doi: 10.1136/bmjopen-2021-050469.
- Glatman-Freedman A, Kaufman Z, Applbaum Y, et al. Respiratory syncytial virus hospitalization burden: a nation-wide population-based analysis, 2000-2017. J Infect. 2020;81(2):297–303. doi: 10.1016/j.jinf.2020.05.078.
- Gunn JS, Marshall JM, Baker S, et al. Salmonella chronic carriage: epidemiology, diagnosis, and gallbladder persistence. Trends Microbiol. 2014;22(11):648–655. doi: 10.1016/j.tim.2014.06.007.
- Lee G, Pan W, Peñataro Yori P, et al. Symptomatic and asymptomatic Campylobacter infections associated with reduced growth in Peruvian children. PLoS Negl Trop Dis. 2013;7(1):e2036. doi: 10.1371/journal.pntd.0002036.
- Allen H, Mitchell HD, Simms I, et al. Evidence for re-infection and persistent carriage of shigella species in adult males reporting domestically acquired infection in England. Clin Microbiol Infect. 2021;27(1):126.e7–126.e13. doi: 10.1016/j.cmi.2020.03.036.
- Wohlwend N, Tiermann S, Risch L, et al. Evaluation of a multiplex real-time PCR assay for detecting major bacterial enteric pathogens in fecal specimens: intestinal inflammation and bacterial load are correlated in campylobacter infections. J Clin Microbiol. 2016;54(9):2262–2266. doi: 10.1128/JCM.00558-16.
- Cybulski RJ, Bateman AC, Bourassa L, et al. Clinical impact of a multiplex gastrointestinal polymerase chain reaction panel in patients with acute gastroenteritis. Clin Infect Dis. 2018;67:1697–1704.
- Gu W, Dutta V, Patrick M, et al. Statistical adjustment of culture-independent diagnostic tests for trend analysis in the foodborne diseases active surveillance network (FoodNet. USA. Int J Epidemiol. 2018;47:1613–1622.

