ABSTRACT
A technological revolution provides nations with access to unprecedented quantities of molecular information, and this is particularly evident in the vast and yet poorly understood realm of the microbiome. Traditionally, many developing countries in Asia, Africa, and South America remain marginal participants in the global flow of biodata, which will eventually affect their productivity and economies. Here, we present the Ecuadorian Microbiome Project (EcuMP) as an integrative initiative to close the research gap in the microbiome for Ecuador. We discuss the relevance that the study of the microbiome has for our understanding of diversity and new forms of production and biocapital. We also evaluate the state of research in metagenomics and the microbiome for South America, with emphasis on Ecuador as a small but biodiverse country. In the strict sense of access, understanding, and technological innovation based on molecular data, we propose the definition of bioliteracy. As indirect estimates of bioliteracy, we measured the number of indexed publications, BioProjects, monthly global internet traffic to GenBank, and patent applications in Espacenet. South America has a notable unevenness in scientific productivity related to the microbiome and metagenomics. Brazil leads productivity, with most of the measured parameters remaining one order of magnitude higher than other countries in the region. Participation of South American countries in the global flow of genomic information dwarfs when compared to the US. To reduce the effects of technological dependency and the associated lack of economic productivity, Ecuador should address the technological gap in the study of the microbiome. Our assessment reveals the urgency to translate the study of microbiomes into a source of technological prowess and the basis for local biocapitals.
Introduction
The unseen biodiversity and its relevance to bioliteracy and biocapital
The estimated number of microorganisms on the planet (i.e. 5 × 1030–10 × 1030) [Citation1,Citation2] is larger than the number of stars in the known universe (1 × 1024) [Citation3]. Despite representing up to 19% of Earth’s biomass or between 1016 and 1017 g, our knowledge of this vast component of biological diversity is still wanting [Citation4]. Some have suggested that at least half of the planet’s biomass belongs to the microbial realm [Citation1,Citation5]. Access to genomic information has become increasingly easier due to the availability of powerful technologies, thus resulting in the discovery of a new realm of culture-independent lineages. These poorly known lineages are often known as the “rare biosphere” or the “dark matter” of microbial diversity [Citation6]. Bacterial lineages without culture isolates consist of most of life’s diversity and some may even represent a new domain of life [Citation7]. This occult and unexplored diversity, which is present in natural environments, in the soil, and water can be a source of natural products with agricultural and biotechnological applications [Citation8]. Thus, the importance to know the microbial universe has become essential to the development of areas such as medicine, agriculture, industry, chemical sciences, and others [Citation9]. This is what we herein call the unseen biodiversity, which holds special relevance to bioliteracy and biocapital [Citation10,Citation11].
Bioliteracy is a term originally coined by Daniel Janzen to express his concern for the accelerated decline of humanity’s knowledge and attention for biodiversity, especially circumscribed to the Barcoding of Life Initiative [Citation11,Citation12]. Janzen’s proposal has been used to denounce our ignorance of nature, in that we only have cataloged a small proportion of the species on Earth [Citation13]. However, some have challenged Janzen’s proposal that the public is “bioilliterate” [Citation13]; particularly from the point of view of the social sciences that propose non-technological forms in which nature and society establish cultural connections [Citation14]. Within the discussion of technological innovation, Enriquez and Martinez [Citation15] have elaborated on the concept of bioliteracy, to stress the importance of understanding and using the molecular resources of biodiversity for new forms of biocapital. It is on this last use of the concept “bioliteracy” that we develop our discussion.
Metagenomic information is an important source of big data for the biological sciences [Citation16]. The capacity to produce, access, and use molecular information has been associated with the level of social and economic development of nations [Citation17], and their possibility for technological innovation [Citation15,Citation18,Citation19]. In a pioneering study, with a methodological approach that has not been replicated to date, Enriquez and Martínez [Citation15] assessed the amount of biodata flow across countries during a period of six months by measuring the number of bytes that were downloaded from GenBank, EMBL (European Molecular Biology Laboratory) and DDBJ (DNA Data Bank of Japan). They showed a significant gap that separated developed nations from regions such as South America. Nearly 92% of all information downloaded from molecular data banks reached the US and nine countries in Europe; with less than 1% being transferred to countries in Asia, Africa and South America. According to Enriquez and Martínez [Citation15], bioliteracy is a necessary condition for new forms of economical development such as a bio-based economy. More recently, Xia and Liu [Citation17] defined the difference between developing and developed countries in the usage patterns of genomic data as a “divide”, with an “overwhelming silence” in the academic community of “third-world countries”.
The establishment and continuous improvement of new technologies that allow the production of genomic information, and its direct application to “profit-oriented enterprise”, “product-making” and “profit-seeking” has led to new concepts in economic theory, such as “biocapital” [Citation10]. Biocapital refers to all biological resources, especially molecules and genomes, that can be transformed into informatic commodities, and that are used to generate value, profit, wealth, and new markets [Citation20]. The biocapital paradigm has been applied to the realm of the microbiome, where genomics has made important progress in the design of “microbial factories” to produce drug precursors, chemical compounds, and plastic monomers [Citation21–23]. However, some believe that the biocapital will have far-reaching effects on current global production systems [Citation24]. It is therefore important for developing countries to consider their role in the global production of information related to what can be considered as bioinformatic commodities [Citation25].
Microbial genomic research in Latin America
South American countries such as Argentina, Brazil, and Venezuela have joined The Earth Microbiome Project (EMP) ([Citation26–28]. The EMP (www.earthmicrobiome.org) is a multidisciplinary initiative that proposes to survey microbial communities at a planetary scale. Another important global initiative is The Terragenome Project, which is an International Soil Metagenome Sequencing Consortium that seeks to complete the sequencing of soil metagenome at a planetary scale [Citation29]. Ecuador has no current participation as an active and contributing member in either of the previously mentioned projects. Ecuador and other countries in South America, with their distinctive geographic and biological diversity, must get involved in these global projects. Refraining from participating contributes to the technological gap that affects their social and economic development as nations.
In an assessment of the status and development of microbiome research in Brazil, Pylro and Roesch [Citation30] established that it has been for the most part neglected, and that research in the country is scattered among numerous small research groups [Citation30]. They highlight biodiversity indexes that place Brazil as one of the most biodiverse countries in the world but remark that microbial diversity is consistently neglected. Recognizing microbes as a central resource for Brazil’s technological and economic development, Pylro and Roesch made a plead to strengthen and unify efforts across the country [Citation30]. Two such efforts have been the creation of the Brazilian Microbiome Project (BMP) and the Brazilian Institute of Science and Technology for Microbiome Studies (INCT), which strive to organize microbiome research at the national level and to integrate with international initiatives. As Brazil, Ecuador is also a country that leads global biodiversity lists and is considered a hotspot of biodiversity [Citation31]; however, its contributions to the study of microbial diversity are considerably smaller [Citation32]. To reduce the scientific and technological gap in the study of microbiomes in Ecuador, the authors of the present study have established an interinstitutional research group called “The Ecuadorian Microbiome Project” (EcuMP). This is a budding effort to survey the unseen diversity in Ecuador’s bacterial ecosystems.
The objective of this study was to analyze the current state of Ecuador’s research in metagenomics and the microbiome compared to all countries in South America. Herein, we discuss the research progress in Ecuador, the potential contribution made by the EcuMP, and the implications that Ecuador’s current state of development in metagenomics and microbiome research has for technological biocapital and bioliteracy.
Methods
We evaluated the state of research in metagenomics and the microbiome for all countries or territories in the South American continent through the PubMed Advanced Search Builder (ASB). The countries considered for analysis were Argentina, Bolivia, Brazil, Chile, Colombia, Ecuador, Guyana, Paraguay, Peru, Surinam, Uruguay, and Venezuela. We also included French Guiana as a territory that is part of the South American continent. PubMed is the search engine for the MEDLINE database and is also part of the Entrez database system [Citation33]. The MEDLINE database includes 200 journals that belong to the regional index Latindex. To include all possible adjectival and demonymic forms for countries, only the root of the country name was used and followed by the wildcard “*”. The search keywords “Ecuador*” and “metagenom*” were applied to all fields of the ASB to assess the number of publications including these words. To provide a comparison basis for Ecuador, a similar search was done for all thirteen countries and territories in the South American continent (e.g. “Argentin*” and “metagenom*”). A similar search was made for studies on the microbiome by using the search term “microbiome*”.
A subsequent advanced Entrez text search in the BioProjects database in GenBank was used to determine the number of matches for the words “Ecuador*”, “Argentin*”, “Venezuela*”, “Brazil*” and the rest of the countries and territories in South America within the “Description” field. The search results in the Entrez system provided a classification within taxonomical groups and research methods, which included the categories “bacteria” and “metagenomes” correspondingly; the reported frequencies or counts in both categories were used as the focus of our analysis. We present the potential limitations of this search approach in the discussion section.
To obtain a rough and indirect estimate of monthly access rates to the NCBI portal (ncbi.nlm.nih.gov), we used the online intelligence tool SimilarWeb [Citation34]. Estimates were obtained between May and July 2018. SimilarWeb is a commonly used traffic analysis tool to perform research and generate insights on estimated online traffic, and it relies on over 50 million users and 1.1 billion records of website visits to infer statistics [Citation35–37]. The rough estimate provided by our traffic measurement approach to the NCBI portal could be used as an indirect metric of bioliteracy (sensu Enriquez & Martínez [Citation15]) or technology transfer (sensu Wang et al. [Citation38]) and an assessment of global participation in the microbiome and metagenomics research. All previously described searches were made on January 16th, 2019.
As an indirect tool to measure internet traffic, caution is required when working with SimilarWeb as it may provide broad estimates only [Citation39,Citation40] and should not be used as a replacement to server monitoring services and log file analyzers when server access is available [Citation41]. However, on servers that receive frequent daily visits by specific countries, such as NCBI and the US, and where large contrasts may be available for other regions of the world, such as Ecuador, results obtained by tools such as SimilarWeb are valuable references that provide a preliminary sense of how different countries may be using resources and data [Citation42].
Metagenomics is a tool for the discovery of new molecules with diverse functions to industry [Citation43] and therefore a potential source of innovation and patenting processes as precursors of biocapital [Citation44]. Patent production does not relate linearly to scientific research [Citation45], but the former is a reliable indicator of the potential that societies have for innovation and the creation of new knowledge [Citation46–48]. To assess the participation of South America in patent creation, we used the advanced search engine available in Espacenet (European Patent Office EPO, worldwide.espacenet.com) [Citation49]. The search, in both title and abstract, was based on the keywords “metagenomic*”, “microbiome”, and “bacteria”, and were individually combined with the name of the applicant country. Espacenet has worldwide coverage and offers information on inventions and technical development from 1836 to the present, with daily coverage. As applicants often file patent applications for the same invention to multiple patent offices, Espacenet consolidates multiple sources and provides access to reference documentation. For comparison of South America as a whole, we also included metrics for the United States as a leading country in the production and use of information related to the microbiome. Ecuador is part of those jurisdictions around the world where organisms are not patentable; however, our search intended to find those technological products that are derived from the study of organisms, such as new methods and processes or synthetic molecules.
To understand the content of the publications found for Ecuador, research trends in the country, and their significance to environmental microbial biodiversity, we applied language analysis. First, publications found in the ASB search were categorized into Scopus subject areas. Second, a word cloud analysis provided a visualization of the most frequent words and a description of the essential information produced for Ecuador. Abstracts of the publications in the four most frequent Scopus categories (i.e. molecular biology, agricultural and biological sciences, immunology and microbiology, and environmental science) were used as the source data for word clouds. The commonality word cloud depicted the most frequent 30 words in a corpus of publications across Scopus categories. The comparison word cloud contrasted word frequencies across documents and provided a depiction of the first 120 words that contributed the most to differentiate among Scopus categories [Citation50]. The word cloud analysis was implemented through the “tm” [Citation51] and “wordcloud” [Citation50] packages in R [Citation52]. For those cases in which Scopus provided more than one category, the authors selected the most appropriate one according to manuscript content and used it for the comparison cloud. Database searches included all available records with no restrictions to time periods. A glossary of terms, with core concepts used in this manuscript, can be found as supplemental information (10.1080/23766808.2021.1938900SI – Glossary of terms).
Results
Comparative analysis of research productivity and publications
A total of fourteen indexed publications were found for Ecuador in the ASB at PubMed for research in metagenomics. Of these, twelve were research-based studies and two reviews. Eleven belonged to studies based on samples obtained from Ecuador’s territory. Three studies were based on research exclusively developed in Ecuadorian territory, while most of the research activities, laboratory procedures, and data analysis for the remaining eleven studies were conducted abroad (). Of these fourteen publications, nine included an author who belongs to an Ecuadorian institution, and five were published exclusively by foreign institutions. Only two had an Ecuadorian researcher as the first author, and three included an Ecuadorian last author. The fourteen publications found through the search included ten with an explicit metagenomic application in the methods and results sections and four that did not include metagenomic approaches or methods ().
Figure 1. Characteristics of publications in Ecuador associated with metagenomics. In the 14 publications found, there is limited participation of local researchers; more noticeably is the large asymmetry between the origin of source samples and the application of research, which is often done overseas
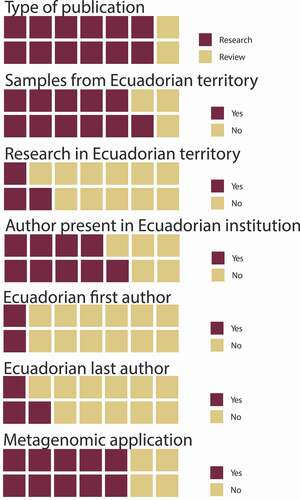
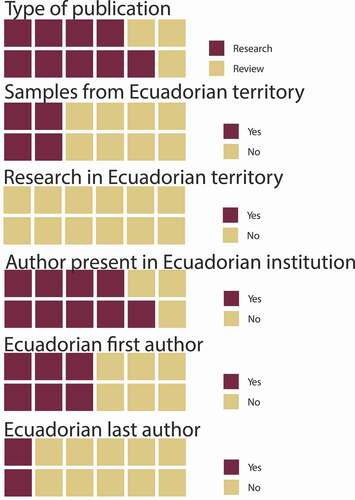
A total of fifteen indexed publications concerning the microbiome were found for Ecuador in the ASB at PubMed: however, three were already included in the metagenomics group and were not considered for the following tally. Three of the twelve publications for the microbiome were reviews and the remaining nine were research-based studies. Four publications included samples from Ecuadorian territory and not one included research done in Ecuador. Nine included an author representing an Ecuadorian institution, six had an Ecuadorian first author, and two had an Ecuadorian last author ().
Figure 2. Characteristics of publications in Ecuador associated with the microbiome. Although with only 12 surveyed publications, there is limited participation of Ecuadorian researchers in the national territory
Both metagenomics and microbiome publications were included in eight Scopus categories, the most frequent being molecular biology, agricultural and biological sciences, and immunology and microbiology. Environmental sciences, a category that should include most of the studies related to microbial ecology, was the fourth most frequent. A single publication was found for the categories of chemical engineering, neuroscience, and chemistry ().
Figure 3. Language analysis through categorization and word clouds of the publications in Ecuador. a) Most studies are associated with the PubMed categories of molecular biology, agricultural and biological sciences, immunology and microbiology, and environmental science. b) The comparison cloud shows how the analyzed publications in the four most frequent PubMed categories differentiate in their use of concepts. c) The commonality cloud is a representation of the most frequent concepts used in all the surveyed publications
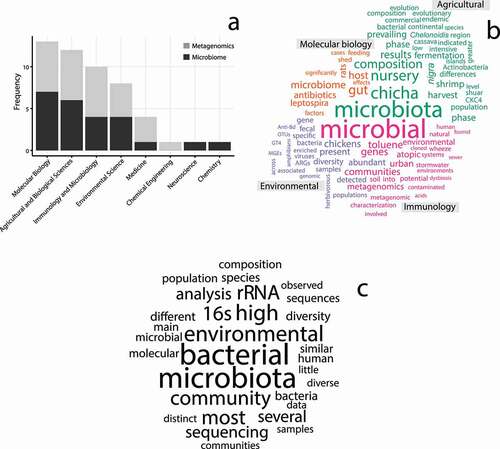
The comparison cloud places heavier weights (i.e. larger words) on those words that contribute the most to differentiate among categories; therefore, the following description is an overview of the central content of the publications on metagenomics and the microbiome for Ecuador () within each Scopus category. The molecular biology category is characterized by studies related to the effect of antibiotics in the microbiome of rat guts and the microbiome associated with the Leptospira infectious cycle. The agricultural and biological sciences category is distinguished by studies on shrimp microbiota (in which the CKC4 bacterial lineage is studied), and the cassava fermentation process that is involved in the production of the indigenous chicha drink. For the immunology and microbiology category, the differentiating trait of the publications is the characterization of microbial communities through metagenomic applications with interest to human and environmental health, such as those associated with the metabolism of hydrocarbons (e.g. toluene). Finally, for the environmental science category, the distinctive concepts are for antibiotic resistance in poultry (including the presence of antibiotic resistance genes (ARGs), and mobile genetic elements (MGEs)), the virome of urban streams affected by fecal contamination, and the distribution of dangerous bacteria to amphibians (). The commonality cloud shows that the publications for Ecuador are often related to the analysis of the microbiota through the sequencing of the 16S rRNA. The composition and diversity of bacterial communities, in a human or environmental context, is often the focus of these studies (). A detailed list of the publications considered in this analysis, including objectives, organisms studied, and main results are included in the supplemental information (101080/23766808.2021.1938900SI – Table 1).
In a regional context, Brazil was the most productive with 375 publications related to the microbiome, followed by Chile (62), Argentina (59), Peru (51), Colombia (42), Venezuela (16), and Ecuador (15). The latter was only more productive when compared to Uruguay and Bolivia with three and four publications each. Brazil led the list of countries in South America with the most publications on metagenomics, totaling 409. Chile was second with 80 publications, followed by Argentina (64) and Peru (64), which together occurred above the third quartile. Both Ecuador and Venezuela were close to the median with 14 and 12 publications respectively. The rest of the countries in South America remained below the first quartile with 8 publications or less ().
Figure 4. Sequence data production and publications on metagenomics and bacteria in South America. Ecuador remains in the median or first quartile for the number of publications and Entrez BioProjects, with either metagenomic information or related to the domain Bacteria
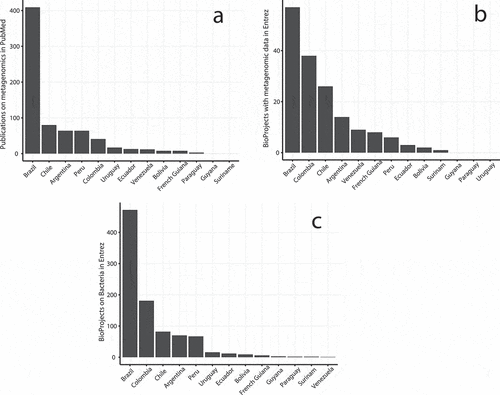
The results of the Entrez text search in the BioProjects database showed that Brazil leads the production of metagenomic data with a total of 57 records. Colombia, Chile, and Argentina remain on or close to the third quartile. Venezuela, French Guiana, and Peru remain close to the median. The rest of the countries, including Ecuador, have three or fewer records and remain on or below the lower quartile (). A similar search for the United States (US) revealed a difference by two orders of magnitude with Brazil, with a total of 3,519 BioProjects associated with the metagenome.
The same Entrez search provided how many projects are related to the domain Bacteria as an organism group. Brazil leads the list with 472 BioProjects in Bacteria, followed by Colombia (181) and Chile (82) and all are above the third quartile. The distribution of BioProjects in Bacteria, as the former two cases ( and ) is highly skewed to the left, where Argentina and Peru, with 70 and 67 records respectively, represent the median and are followed by Uruguay with 16 records. Ecuador, with 12 records, represents the first quartile and the remaining countries have nine or fewer records (). The difference between Brazil and the US is minor in this case, the latter having a total of 339 BioProjects, slightly less than the former.
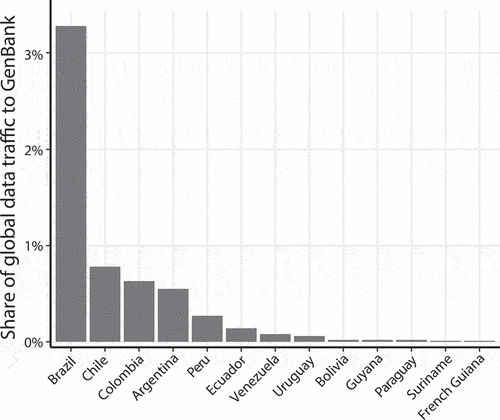
Access to NCBI is an order of magnitude larger for Brazil when compared with the rest of South American countries, with over 3% of the global traffic share. Ecuador remains close to the median and leads the tail of countries with less than 0.1% of global participation in accessing the information on NCBI (). The United States is an order of magnitude larger than Brazil, with 33.53% of the total share of monthly traffic to NCBI. No country in South America registered patent applications with words related to metagenomics or the microbiome. Only Chile (17), Brazil (7), and Colombia (4) registered patent applications that included the keyword bacteria. There is a stark difference when South America is compared to the US. The latter has 828 patent applications related to bacteria, 7 to metagenomics, and 3 to the microbiome. A worldwide search revealed 159 applications with words related to metagenomics.
Figure 5. A rough estimate of access to the NCBI main portal as a percentage of global traffic share between May and July 2018 (three months). A sharp contrast is present between Brazil and the rest of the countries in South America. Ecuador leads the tail of countries with less than 0.1% of global participation
Conclusions and discussion
Research production and publications
The present study presents an analysis of scientific productivity by South American countries in both the microbiome and metagenomic fields. From the assessment of the evidence available in international public databases, such as PubMed, Entrez, and Espacenet, it is evident that South America suffers from a pronounced unevenness, with Brazil as the single most important center of research in the region. As has been hinted in the present study, when metrics for the US are included in the comparisons, the gap in knowledge production increases to an order of magnitude larger than Brazil, the latter as the leading country in the South American region.
The role of Chile, as the leading nation in the region for patents on bacteria, may respond to processes within the country that are not necessarily related to the measurements of scientific productivity we used in this study (e.g. number of publications); particularly, since there are very small numbers to compare with for other countries in South America, we may see this extreme value for Chile as the result of unique circumstances that provide a remarkable difference to the overall tendencies of countries within the region. This is particularly true if we consider the non-linearity by which patents may be produced and that could be decoupled from the processes and conditions involved in the production of scientific literature and academic knowledge [Citation45,Citation53]. What drives Chile as the leading nation in the region for patents on bacteria requires analyses and answers that remain beyond the scope of the current study. Further exploration is needed to compare Chile’s drivers for innovation in the microbiological field with the conditions for other countries in South America; especially as the innovation environments and incentives do not necessarily perform similarly across countries and societies [Citation54]. How countries produce and file patents may depend on numerous and varied factors, from macroeconomic processes by which industry and academy are financed and taxed, including lines of investment and capital risk, to micromanagement factors such as patent filing procedures and bureaucratic requirements [Citation55]. Cultural values, human capital, trust, infrastructure, and laws are also relevant factors involved in the production of patents and other forms of innovation [Citation46,Citation48]. Our measurement of patents is basic and serves only as a very preliminary insight into a more complex system, which requires measurement and explanation.
Of relevance is the extremely limited participation of South America in the data transfer to NCBI, where the region essentially remains secluded from the global flow of genomic information. This agrees with the previous findings by Enriquez and Martínez [Citation15] and Xia and Liu [Citation17]. However, as we did not have access to the NCBI servers for direct measurement of traffic, our indirect approach is only approximate, limited in its scope, and cannot be strictly compared to dedicated software installed at host servers for traffic monitoring [Citation39,Citation40]. The very small number of patent applications for the region may be a symptom of the necessity for new strategies to strengthen bioliteracy and research on the microbiome and its associated genomes.
Research trends in Ecuador
The studies in Ecuador on metagenomics and the microbiome are mostly in aspects related to general molecular biology applications, agricultural sciences, public health, and to a lesser extent in environmental sciences. For a matrix detailing the assessed publications, please refer to 101080/23766808.2021.1938900SI – Table 1. Studies related to direct applications in conservation, such as the study of water quality or pathogens affecting amphibian populations (10.1080/23766808.2021.1938900SI – Table 1), are infrequent as most have applied interests to public health (gut microbiome, hydrocarbon degradation) and industrial productivity (e.g. poultry or shrimp). Within the reduced corpus of publications for Ecuador, there is a clear lack of efforts in the field of microbial ecology through powerful methods such as metagenomics. No studies on soil metagenomic diversity have been published for Ecuador (10.1080/23766808.2021.1938900SI – Table 1).
The Ecuadorian Microbiome Project started in 2016, with the ambitious goal to catalog the bacterial microbial diversity in Ecuador. The EcuMP follows the methods proposed by the Earth Microbiome Project (www.earthmicrobiome.org/protocols-and-standards/) [Citation27] and minor modifications to the 16S Illumina amplicon protocol for samples from extreme environments [Citation56]. Metagenomic sequencing has been done exclusively on the MiSeq Illumina platform. The application of these standardized methods for data acquisition and analysis allows the comparison of Ecuadorian bacterial microbiomes to others from around the world. For all samples, we also record physicochemical variables, which provide a basic ecological framework to understand the causes or effects of microbial diversity (e.g. electrical conductivity, content of organic matter, humidity, cationic exchange, dissolved oxygen, pH, temperature, and presence of P, N, Ca, Mg, SO4). We encourage the standardization of the methods used in the EcuMP to coordinate complementary efforts by other research groups in the country and contribute to a catalog of Ecuadorian microbiomes.
The EcuMP has collected and processed bacterial microbiome samples from over 1,000 sites, including tropical rainforest affected by different intensities of human activity, cloud forest, dry forest, Andean-Choco forest, and extreme environments including hot springs, Andean volcanoes and glaciers, and deep-sea muds. Immediately after collection, the total DNA for each sample was isolated. Metagenomic sequences are available for 500 samples for a total of 24,585,878 giga base pairs, 81,680,662 sequences, and 2.72 Terabytes of data. For taxonomic assignment, biodiversity, and phylogenetic analysis we currently use bioinformatic tools, including dada2 [Citation57], QIIME 1 [Citation58], QIIME 2 [Citation59], and Mothur [Citation60]. Physicochemical data is used for numerical ecology to understand factors driving microbiome biodiversity patterns. Preliminary results are extensive, and a broad snapshot of Ecuador’s biodiverse microbiome is being described and will be published during 2021.
Although the EcuMP remains far from what the BMP has accomplished in Brazil, we have gathered hundreds of samples and metagenomic data from a wide range of habitats, elevations, and regions, including tropical forests and extreme environments such as Andean volcanoes. The EcuMP will soon contribute to new insights into understanding the range of variability and structure of bacterial communities in Ecuador. Such publications will enable the EcuMP to establish links of collaboration with regional and international partners. Further stages of the EcuMP will consider the role of climate change and other major abiotic and biotic components associated with biological diversity. The evaluation of functional diversity and the exploration of the microbial transcriptome, proteome, and metabolome are considered long-term goals of the EcuMP and expected within a segment of five years of uninterrupted development of the EcuMP in Ecuador.
Our data will only have relevance if we share it with the global community to contribute to knowledge discovery and innovation. Data management in the EcuMP project pays special attention to the principles of findability, accessibility, interoperability, and reusability (FAIR) [Citation61]. The application of the FAIR principles to the data gathered by the EcuMP will contribute to strengthen further efforts in understanding the microbiome and guarantee that our data becomes reusable research objects, and acquire as much social and scientific value as possible [Citation62].
Potential sources of bias and future improvements
The use of keywords in the previously described searches provided an indirect measurement of how active countries are in the different categories that were considered. However, the results of these searches do not specify if core research and data production was made within the country or occurred elsewhere. In most cases, search results do not inform us if samples were obtained from the source country and taken abroad for analytical processes or if there was at least a minimum of collaboration within the country to produce publications and sequence data. To understand to what degree researchers, research centers, and other institutions within a country actively participate and contribute to microbiome and metagenomic research will require a finer assessment of the details in each recorded publication and registry in the assessed databases.
Many biodiverse countries are part of the international agreement known as the Nagoya Protocol, which proposes a general mechanism for fair and equitable sharing of benefits arising from research and technological innovation on genetic resources. Ecuador has developed a corpus of legislation in concordance to the Nagoya Protocol, a specific component of which is the enforced participation of local researchers in international research initiatives related to genetic resources [Citation63]. It remains to be assessed the extent to which the analyzed publications have been the product of this enforced mechanism of collaboration, and if the expected transfer of knowledge and technology has been achieved.
Another potential bias in our sampling approach is multiple submissions within a project by a single research group, laboratory, or research institute. This will overestimate how much a country invests in metagenomic and microbiome research. An instance of such a case is Venezuela, where eight of the nine metagenome BioProjects have been submitted by a single project from one research institute. Nevertheless, our study provides a general overview of how countries in the South American region participate in the production of microbiome and metagenomic knowledge.
Challenges and prospects
Most developing countries have recognized the importance of microbial metagenomic research, with the establishment of well-funded national programs [Citation64]. Setting aside the production from developed regions around the world, there is a substantial gap in the production of metagenomic and microbiome knowledge within South America. When Ecuador is compared to Brazil, Argentina, Chile, and Colombia, such a gap consists of at least an order of magnitude in the production of metagenomic information. Ecuador consistently lags behind the latter countries, and although it remains in the second or third quartiles, it leads the group of countries with the weakest production. Brazil is remarkable and stands out far beyond what any other country has produced in terms of publications and metagenomic data in South America.
The literature associated with Ecuador is mostly foreign production with only secondary participation by Ecuadorian authors. Independently of the origin of the data and samples, little to no processes, analyses, and technological activities occur within the Ecuadorian territory. The marginal role that Ecuadorian institutions and accompanying authors have in studies related to bacterial metagenomics or the microbiome in Ecuador is both a consequence and a cause of the technological gap that is observed within the South American region.
It may be argued that the differences in scientific productivity presented in this study are well-known facts, which cut across all disciplines and fields of knowledge. However, we argue, such facts are circumscribed to a small community of science specialists and the discussion does not reach into the broader decision-making sector. We believe that a critical assessment of the regional situation of Ecuador in relevant areas of scientific and technological importance is necessary for establishing sound national strategies, especially when this information becomes available to broader audiences.
It is a well-known fact that researchers in developing countries experience a range of social, cultural, and economic deterrents and pitfalls for the publication of their work in international journals [Citation65–67]. Language is one of the main barriers involved in the divide between mainstream science and local publications or even the presence of forgotten studies that remain unpublished [Citation68]; especially when we consider that English is the predominant lingua franca among scientific journals. The coverage of the literature quantified in the present study is restricted mostly to MEDLINE as the primary bibliographic database source for PubMed. The extent to which native South American research on the microbiome and metagenomics is not properly represented in the assessed databases remains unknown to us. Factors such as limitations to communicate in written English, access to competitive technology, political instability, and a set of cumbersome laws and regulations to access and work with genetic assets may all interact together for the observed lack of productivity in the region [Citation69–71]. The application of the Nagoya Protocol, of which Ecuador is signatory through local legislation has been extensively criticized by the scientific communities of many countries, due mainly to its complexity and excessive requirements [Citation63,Citation72–74]. In this sense, the contrasting differences between Brazil and the rest of South American countries, or the case of Chile for patents on bacteria, should be studied in additional detail to understand those social, cultural, and economic factors that are most favorable to the study of the microbiome and scientific research within the South American region.
Patents are the result of complex social and cultural interactions and are one “core artifact” in a broader system by which a bioeconomy may develop [Citation75]. Thus, the term “bioeconomy”, in its broadest sense, consists of the transformation of natural resources to achieve food security, sustainability, avoidance of fossil-fuel dependency, climate change mitigation, strengthening of the job market, and rural development. A range of strategies may be applied to reach many forms of bioeconomies, depending on the nature of a country’s geography, culture, and state of development, from diversified high-tech to basic-primary economies [Citation76]. The many ways and strategies a country may transform its production matrix into a new bioeconomy [Citation77] must include not only the promotion of new forms of technology and knowledge (such as patents and scientific publications) but also new forms of political visions and social interactions [Citation75,Citation78,Citation79].
Besides cumbersome laws and language barriers; the development of microbial ecology, as a discipline in Ecuador, shares with all fields of the natural sciences and biotechnology other critical obstacles. These include: 1) excessive costs and taxes for equipment and supplies, most of which have to be imported through intermediaries along extended periods, 2) long-term research usually lacks long-term funding to transpire into internationally relevant initiatives, 3) there is no clear separation between transient political moments and sustained national strategies in science and technology (sensu Bush [Citation80]), and 4), universities in Ecuador have been traditionally disconnected from scientific inquiry and committed to replicating knowledge produced elsewhere [Citation70].
A thorough assessment of obstacles for research in microbial biodiversity goes beyond the scope of this manuscript. We consider that the seminal proposal by Bush [Citation80], for the successful science and technology system implemented in the US, could provide hints for a sound strategy in Ecuador. Particularly important among Bush’s proposal is the principle of independence and stability that long-term research requires in both funding and political interests and decisions. Open markets with zero tax liability for scientific equipment and supplies, relaxed controls for public funding spending in science and technology, and a stable long-term national fund for developing extended science and technology programs are all part of what we believe is necessary to remove the discussed obstacles.
A synopsis of the 787 accredited researchers by the Ecuadorian national authority of higher education, science, technology, and innovation (SENESCYT), reveals that only 4 are related to research in microbial diversity, metagenomics, and the microbiome. The Constitution of Ecuador determines that 0.55% of the national GDP must be allocated to national scientific research activities. According to the World Development Indicators database by the World Bank and reported in Roser and Ortiz-Ospina [Citation81], the most recent report for Ecuador is from 2014 and corresponds to 0.44% of the GDP. This is larger when compared to Colombia (0.30%) and Peru (0.11%), but considerably smaller than the investment in Brazil (1.17%). Reported expenditures to the World Bank do not necessarily reflect direct investments into research programs, with some arguing that the effective investment for scientific research and development is close to 1% of that required by the Constitution [Citation82]. Given the small available workforce dedicated to studying microbial diversity in Ecuador, and that our national expenditure in science and technology is reduced, at least in terms of direct financing to research programs [Citation82], a national strategy is required for the continued collaboration of Ecuadorian institutions with scientists from developed countries. Genomic research is relatively expensive and international collaboration is essential for strengthening national competitiveness.
To reduce the breach within the South American region in the production of metagenomic and microbiome knowledge, efforts such as the Ecuadorian Microbiome Project require steady support of government agencies related to scientific and technological development. Steadiness in budget provisions and independence from the transitory political periods have both been a foundational component of the United States strategy for science and technology, since the publication of its first national scientific program [Citation80]. The simplification and rationalization of the current national regulations and processes for the procurement of imported technology and machines are also essential. It is particularly critical to the success of bioprospecting projects that the processes for obtaining permits and contracts to access biological samples and information get streamlined; or otherwise, these bureaucratic establishments will remain unsurpassable obstacles to development [Citation83–84]. The national strategies to quantify and valorize biological diversity cannot any longer neglect the important realm of microorganisms, on which most of the current and future biotechnologies depend. We make a call for local teams in Ecuador and abroad, that are involved in research on microbial diversity, to join the Ecuadorian Microbiome Project and help strengthen this important area of scientific and technological development.
Technological progress towards an economy based on biocapitals is especially crucial for developing countries that depend on non-renewable commodities such as petroleum and raw agricultural materials with little added technological value. How nations assign value to their biological resources, such as those held in the microbiome, is particularly critical nowadays in the age of rapidly expanding technologies that can read, design, and translate the digital language of life. The economic welfare of nations may increasingly come to depend on how rapidly they adapt to new paradigms of technological biocapitals and bioliteracy. Modern biotechnology is at the heart of political, economic, and social debates, both internationally and nationally. The state of microbiome and metagenomic research in Ecuador and the South American region remains to be developed, such that the observed gap will no longer represent a risk to technological independence and economic productivity.
Disclosure of potential conflicts of interest
The authors have not received any direct funding or financial support for the publication of this review. The authors do not hold any conflict-of-interest with the entities, organizations, or groups mentioned in the present review. This manuscript is the result of shared academic collaboration by the authors as a research group and is not the product of writing assistance by third parties or external associates to the research group; the latter is the sole responsible for the authorship of the present review.
Geolocation information
We place as a reference for geolocation the research lab at the Institute of Research on Zoonoses (CIZ), −0.199303, −78.505963.
Supplemental Material
Download Zip (40.1 KB)Acknowledgments
Our project has been funded by the Research Grants Programme of The World Academy of Sciences (TWAS), to which we extend our gratitude. Our project has also received funding from the Academy of Research and Higher Education (ARES) of the Belgian Government, and as part of an integrated collaboration program with Universidad Central del Ecuador. The Ecuadorian Microbiome Project would not have been possible without the support of our home institutions, Central University of Ecuador and the Technical University of the North to which we extend our gratitude. The authors are grateful to Dr. Spiros Agathos of Université Catholique de Louvain and to Dr. Félix Sangari of the University of Cantabria for their constant support and valuable advice. Our gratitude is extended to three anonymous reviewers, whose comments and suggestions contributed to the improvement of our manuscript.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci U S A. 1998;95(12):6578–6583.
- Wooley JC, Godzik A, Friedberg I. A primer on metagenomics. PLoS Comput Biol. 2010;6(2):e1000667.
- van Dokkum PG, Conroy C. A substantial population of low-mass stars in luminous elliptical galaxies. Nature. 2010;468(7326):940.
- McMahon S, Parnell J. Weighing the deep continental biosphere. FEMS Microbiol Ecol. 2014;87(1):113–120.
- Robbins RJ, Krishtalka L, Wooley JC. Advances in biodiversity: metagenomics and the unveiling of biological dark matter. Stand Genomic Sci. 2016;11(1):69.
- Rinke C, Schwientek P, Sczyrba A, et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499(7459):431.
- Hug LA, Baker BJ, Anantharaman K, et al. A new view of the tree of life. Nat Microbiol. 2016;1(5):16048.
- Gillespie DE, Rondon MR, Williamson LLHJ. Metagenomic libraries from uncultured microorganisms. In: Osborn AMSC, editor. Mol. Microb. Ecol. New York: Taylor & Francis; 2005. p. 261–279.
- Timmis K, de Vos WM, Ramos JL, et al. The contribution of microbial biotechnology to sustainable development goals. Microbiol Biotechnol. 2017;10(5):984–987.
- Helmreich S. Species of biocapital. Sci Cult. (Lond). 2008;17(4):463–478.
- Janzen DH. Hope for tropical biodiversity through true bioliteracy. Biotropica. 2010;42(5):540–542.
- Janzen DH, Godfray HCJ, Knapp S. Now is the time. Philos Trans R Soc Lond B Biol Sci. 2004;359(1444):731–732.
- Pedroza Flores R, Villalobos Monroy G, Reyes Fabela A. A critique on the concept of social accountability in higher education. J Educ Learn. 2015;4:1–9.
- Ellis R, Waterton C, Wynne B. Taxonomy, biodiversity and their publics in twenty-first-century DNA barcoding. Public Underst. Sci. 2010;19(4):497–512.
- Enriquez J, Martínez R. The biotechonomy (1.0): a rough map of bio-data flow. Harvard Business School Working Paper. 2002;3(28):1–21.
- Vestergaard G, Schulz S, Schöler A, et al. Making big data smart—how to use metagenomics to understand soil quality. Biol Fertil Soils. 2017;53(5):479–484.
- Xia J, Liu Y. Usage patterns of open genomic data. Coll Res Libr. 2013;74(2):195–206.
- Bevan MW, Uauy C, Wulff BBH, et al. Genomic innovation for crop improvement. Nature. 2017;543(7645):346–354.
- Khan R, Mittelman D. Consumer genomics will change your life, whether you get tested or not. Genome Biol. 2018;19(1):120.
- Milne R. From people with dementia to people with data: participation and value in Alzheimer’s disease research. Biosocieties. 2018;13(3):623–639.
- Ibrahim MHA, Lebbe L, Willems A, et al. Chelatococcus thermostellatus sp. nov., a new thermophile for bioplastic synthesis: comparative phylogenetic and physiological study. AMB Express. 2016;6(1):39.
- Mutreja A. Chapter 8 – Microbial genomics: diagnosis, prevention, and treatment. In: Kumar D, Antonarakis SBT-M, Hg, editors. Medical and Health Genomics. Oxford: Academic Press; 2016. p. 101–106.
- Marx V. How some labs put more bio into biomaterials. Nat Methods. 2019;16(5):365–368.
- Priefer C, Jörissen J, Frör O. Pathways to shape the bioeconomy. Resources. 2017;6(10):261–279.
- De Las Rivas J, Bonavides-Martínez C, Campos-Laborie FJ. Bioinformatics in Latin America and SoIBio impact, a tale of spin-off and expansion around genomes and protein structures. Brief Bioinform. 2017;18(6):1091.
- Thompson LR, Sanders JG, McDonald D, et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature. 2017;551(7681):457.
- Gilbert JA, Meyer F, Jansson J, et al. The Earth Microbiome Project: meeting report of the “1(st) EMP meeting on sample selection and acquisition” at Argonne national laboratory. Stand Genomic Sci. 2010;3:249–253.
- Gilbert JA, Jansson JK, Knight R. The Earth Microbiome Project: successes and aspirations. BMC Biol. 2014;12(1):69.
- Vogel TM, Simonet P, Jansson JK, et al. TerraGenome: a consortium for the sequencing of a soil metagenome. Nat Rev Microbiol. 2009;7(4):252.
- Pylro V, Roesch L. The Brazilian Microbiome Project. In: Pylro VRL, editor. The Brazilian Microbiome, Current Status and Perspectives. Cham: Springer; 2017. p. 1–6.
- Montoya E, Keen HF, Luzuriaga CX, et al. Long-term vegetation dynamics in a megadiverse hotspot: the ice-age record of a pre-montane forest of central Ecuador. Front Plant Sci. 2018;9:196.
- Mittermeier R, Robles Gil P, Hoffmann M, et al. Hotspots revisited. Earth’s biologically richest and most endangered terrestrial ecoregions. Conserv Int. Mexico City: CEMEX S. A de C. V; 2004.
- Lu Z. PubMed and beyond: a survey of web tools for searching biomedical literature. Database (Oxford). 2011;2011:baq036.
- SimilarWeb LTD 2019. SimilarWeb.com – competitive Intelligence Tool [Internet]. [ cited 2019 Jan 16]. Available from: http://www.similarweb.com/.
- Ismailova R, Inal Y. Accessibility evaluation of top university websites: a comparative study of Kyrgyzstan, Azerbaijan, Kazakhstan and Turkey. Univers Access Inf Soc. 2018;17(2):437–445.
- Kagan S, Bekkerman R. Predicting purchase behavior of website audiences. Int J Electron. Commer. 2018;22(4):510–539.
- Goncalves JR, de Oliveira DD, Sassi RJ. Classification of web history tools through web analysis. HCI cybersecurity Priv. Trust. Fist Int. Conf. Orlando, FL; 2019. p. 266–276.
- Wang M, Pfleeger S, Adamson D, et al. Technology transfer of federally funded R&D: perspectives from a forum. Santa Monica, CA: RAND Science and Technology Policy Institute; 2003.
- Król K, Halva J. Measuring efficiency of websites of agrotouristic farms from Poland and Slovakia. Econ Reg Stud. 2017;10:50–59.
- David P, Martin P. Website traffic measurement and rankings: competitive intelligence tools examination. Int J Web Inf Syst. 2018;14(4):423–437.
- Mbaye B, Gueye A, Banse D, et al. Africa’s online access: what data is getting accessed and where it is hosted? Innovations and interdisciplinary solutions for underserved areas. Bassioni G, Kebe CMF, Gueye A, et al., editors. Cham: Springer International Publishing; 2019. p.50–61.
- Król K. Marketing potential of websites of rural tourism facilities in Poland. Econ Reg Stud. 2019;12(2):158–172.
- Lorenz P, Eck J. Metagenomics and industrial applications. Nat Rev Microbiol. 2005;3(6):510–516.
- Rajan KS. Biocapital, the constitution of postgenomic life. Durham and London: Duke University Press; 2006.
- Oldham P, Hall S, Forero O. Biological diversity in the patent system. PLoS One. 2013;8(11):e78737–e78737.
- Şener S, Sarıdoğan E. The effects of science-technology-innovation on competitiveness and economic growth. Procedia Soc Behav Sci. 2011;24:815–828.
- Moser P. Patents and innovation: evidence from economic history. J Econ Perspect. 2013;27(1):23–44.
- Zhu B, Habisch A, Thogersen J. The importance of cultural values and trust for innovation — a european study. Int J Innov. Manag. 2017;22(2):1850017.
- Organisation EP Espacenet: patent search [Internet]. Munich: The European Patent Organization; [cited 2019 Mar 25]. Available from: worldwide.espacenet.com.
- Fellows I. wordcloud: Word Clouds. [R package]. Version 2.6. 2018. Available from: https://cran.r-project.org/package=wordcloud.
- Feinerer I, Hornik K. tm: Text Mining Package. [R package]. Version 0.7–8. 2008. Available from: https://r-forge.r-project.org/projects/tm/.
- R Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing [Downloadable software]. Version 3.5.3. Viena: R Foundation for Statistical Computing, Austria; 2020. Available from: https://www.r-project.org/.
- Ribeiro B, Shapira P. Private and public values of innovation: a patent analysis of synthetic biology. Res Policy. 2020;49(1):103875.
- Cohen WM, Goto A, Nagata A, et al. R&D spillovers, patents and the incentives to innovate in Japan and the United States. Res Policy. 2002;31(8–9):1349–1367.
- Ryan JC, Daly TM. Barriers to innovation and knowledge generation: the challenges of conducting business and social research in an emerging country context. J Innov Knowl. 2019;4(1):47–54.
- Tighe S, Afshinnekoo E, Rock TM, et al. Genomic methods and microbiological technologies for profiling novel and extreme environments for the Extreme Microbiome Project (XMP). Journal of Biomolecular Techniques : JBT. 2017;28(1):31–39..
- Callahan BJ, McMurdie PJ, Rosen MJ, et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nature Methods. 2016;13(7):581–583.
- Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336..
- Bolyen E, Rideout JR, Dillon MR, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37(8):852–857.
- Schloss PD, Westcott SL, Ryabin T, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75(23):7537–7541. /10/02. 2009.
- Wilkinson MD, Dumontier M, Aalbersberg IJJ, et al. The FAIR guiding principles for scientific data management and stewardship. Sci Data. 2016;3(1):160018.
- Mons B, Neylon C, Velterop J, et al. Cloudy, increasingly FAIR; revisiting the FAIR Data guiding principles for the European Open Science Cloud. Inf Serv Use. 2017;37:49–56.
- Rojas Blanco DL. Vicisitudes del Protocolo de Nagoya en Colombia. Gestión y Ambiente. 2013;16:17–23.
- Stulberg E, Fravel D, Proctor LM, et al. An assessment of US microbiome research. Nat Microbiol. 2016;1(1):15015.
- Packer A. The SciELO open access: a gold way from the south. Can. J High Educ. 2010;39(3):111–126.
- Clavero M. “Awkward wording. Rephrase”: linguistic injustice in ecological journals. Trends Ecol Evol. 2010;25(10):552–553.
- Salager-Meyer F. Writing and publishing in peripheral scholarly journals: how to enhance the global influence of multilingual scholars? J English Acad Purp. 2014;13:78–82.
- Pronskikh V. Linguistic privilege and justice: what can we learn from STEM? Philos. Pap. 2018;47:71–92.
- Hidalgo CA, Hausmann R. The building blocks of economic complexity. Proc Natl Acad Sci. 2009;106(26):10570.
- Feyen J, Van Hoof HB. An analysis of the relationship between higher education performance and socio-economic and technological indicators: the Latin American case study. MASKANA. 2013;4(2):1–20.
- Magee SP. Lawyers as spam, congressional capture explains why U.S. lawyers exceed the optimum. In: Buckley FH, editor. Am. ilness, essays rule law. London: Yale University Press; 2013. p. 100–117.
- Alves RJ, Weksler M, Oliveira JA, et al. Brazilian legislation on genetic heritage harms Biodiversity Convention goals and threatens basic biology research and education. Anais da Academia Brasileira de Ciencias. 2018;90(2):1279–1284.
- Bockmann FA, Rodrigues MT, Kohsldorf T, et al. Brazil´s government attacks biodiversity. Science. 2018;360(6391):865.
- Prathapan KD, Pethiyagoda R, Bawa KS, et al. When the cure kills—CBD limits biodiversity research. Science. 2018;360(6396):1405–1406.
- Befort N. Going beyond definitions to understand tensions within the bioeconomy: the contribution of sociotechnical regimes to contested fields. Technol Forecast Soc Change. 2020;153:119923.
- Aguilar A, Twardowski T, Wohlgemuth R. Bioeconomy for sustainable development. Biotechnol J. 2019;14(8):1800638.
- Petersen A, Krisjansen I. Assembling ‘the bioeconomy’: exploiting the power of the promissory life sciences. J Sociol. 2015;51(1):28–46.
- Goven J, Pavone V. The bioeconomy as political project: a Polanyian analysis. Sci Technol Hum Values. 2015;40(3):302–337.
- Aguilar A, Wohlgemuth R, Twardowski T. Perspectives on bioeconomy. N Biotechnol. 2018;40:181–184.
- Bush V. Science, the endless frontier: a report to the President on a program for postwar scientific research. Washington DC: United States Government Printing Office; 1945.
- Roser M, Ortiz-Ospina E. Global rise of education [Internet]. OurWorldInData.org. 2019 [cited 2019 Mar 25]. Available from: https://ourworldindata.org/global-rise-of-education.
- Paz y Miño C. Lo inédito de Inédita. Redacción Médica [Internet]. 2018 [cited 2019 Mar 25]. Available from: https://www.edicionmedica.ec/opinion/lo-inedito-de-inedita-2028.
- Fernández F. The impediment for the study of the biodiversity in Colombia. Caldasia. 2011;33:3–5.
- Páez VP. Colombia’s tax on wildlife studies. Science. 2016;354(6309):191.
- Rusch DB, Halpern AL, Sutton G, et al. The Sorcerer II global ocean sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5(3):e77–e77.
- Sharma AK, Zhaxybayeva O, Papke RT, et al. Actinorhodopsins: proteorhodopsin-like gene sequences found predominantly in non-marine environments. Environ Microbiol. 2008;10(4):1039–1056.
- Cardenas PA, Cooper PJ, Cox MJ, et al. Upper airways microbiota in antibiotic-naïve wheezing and healthy infants from the tropics of rural Ecuador. PLoS One. 2012;7(10):e46803–e46803. /10/05. 2012.
- Loire E, Chiari Y, Bernard A, et al. Population genomics of the endangered giant Galápagos tortoise. Genome Biol. 2013;14(12):R136–R136.
- Colehour AM, Meadow JF, Liebert MA, et al. Local domestication of lactic acid bacteria via cassava beer fermentation. PeerJ. 2014;2:e479–e479.
- Hong P-Y, Mao Y, Ortiz-Kofoed S, et al. Metagenomic-based study of the phylogenetic and functional gene diversity in Galápagos land and marine iguanas. Microb Ecol. 2015;69(2):444–456.
- Bresciano JC, Salvador CA, Paz-y-Miño C, et al. Variation in the presence of anti-Batrachochytrium dendrobatidis bacteria of amphibians across life stages and elevations in Ecuador. Ecohealth. 2015;12(2):310–319.
- McLellan SL, Fisher JC, Newton RJ. The microbiome of urban waters. Int Microbiol. 2015;18(3):141–149.
- Trueba AF, Ritz T, Trueba G. The role of the microbiome in the relationship of asthma and affective disorders. In: Lyte M, editor. Microb. Endocrinol. interkingdom Signal Infect Dis Heal. Cham: Springer International Publishing; 2016. p. 263–288.
- Bouhajja E, Agathos SN, George IF. Metagenomics: probing pollutant fate in natural and engineered ecosystems. Biotechnol Adv. 2016;34(8):1413–1426.
- Nesme J, Achouak W, Agathos SN, et al. Back to the future of soil metagenomics. Front Microbiol. 2016;7:73.
- Bouhajja E, McGuire M, Liles MR, et al. Identification of novel toluene monooxygenase genes in a hydrocarbon-polluted sediment using sequence- and function-based screening of metagenomic libraries. Appl Microbiol Biotechnol. 2017;101(2):797–808.
- Arrieta M-C, Arévalo A, Stiemsma L, et al. Associations between infant fungal and bacterial dysbiosis and childhood atopic wheeze in a nonindustrialized setting. J Allergy Clin. Immunol. 2017/ 12/11. 2018;142(2):424–434.e10.
- Barragan V, Olivas S, Keim P, et al. Critical knowledge gaps in our understanding of environmental cycling and transmission of Leptospira spp. Appl Environ Microbiol. 2017;83(19):e01190–17.
- Barragan V, Nieto N, Keim P, et al. Meta-analysis to estimate the load of Leptospira excreted in urine: beyond rats as important sources of transmission in low-income rural communities. BMC Res Notes. 2017;10(1):71.
- Ben-Yosef M, Zaada DSY, Dudaniec RY, et al. Host-specific associations affect the microbiome of Philornis downsi, an introduced parasite to the Galápagos Islands. Mol Ecol. 2017;26(18):4644–4656.
- Pfeiffer S, Mitter B, Oswald A, et al. Rhizosphere microbiomes of potato cultivated in the high Andes show stable and dynamic core microbiomes with different responses to plant development. FEMS Microbiol Ecol. 2016;93(2):93.
- Carvajal-Aldaz DG, Guice JL, Page RC, et al. Simultaneous delivery of antibiotics neomycin and ampicillin in drinking water inhibits fermentation of resistant starch in rats. Mol Nutr Food Res. 2016;61(3):1600609.
- Loayza Villa MF, Herrera Sevilla VL, Vivar-Diaz N. Detection of Helicobacter pylori DNA in formalin-fixed paraffin-embedded gastric biopsies using laser microdissection and qPCR. In: Bishop-Lilly KA, editor. Diagnostic Bacteriol methods Protoc. New York: Springer New York; 2017. p. 71–88.
- Gainza O, Ramírez C, Ramos AS, et al. Intestinal microbiota of white shrimp Penaeus vannamei under intensive cultivation conditions in Ecuador. Microb Ecol. 2018;75(3):562–568.
- Suzuki TA, Martins FM, Nachman MW. Altitudinal variation of the gut microbiota in wild house mice. Mol Ecol. 2019;28(9):2378–2390.
- Burgos FA, Ray CL, Arias CR. Bacterial diversity and community structure of the intestinal microbiome of Channel Catfish (Ictalurus punctatus) during ontogenesis. Syst Appl Microbiol. 2018;41(5):494–505.
- Guo X, Stedtfeld RD, Hedman H, et al. Antibiotic resistome associated with small-scale poultry production in rural Ecuador. Environ Sci Technol. 2018;52(15):8165–8172.
- Wise EL, Pullan ST, Márquez S, et al. Isolation of Oropouche virus from febrile patient, Ecuador. Emerg Infect Dis. 2018;24(5):935–937.
- Santos JC, Tarvin RD, O’Connell LA, et al. Diversity within diversity: parasite species richness in poison frogs assessed by transcriptomics. Mol Phylogenet Evol. 2018;125:40–50.
- Guerrero-Latorre L, Romero B, Bonifaz E, et al. Quito’s virome: metagenomic analysis of viral diversity in urban streams of Ecuador’s capital city. Sci Total Environ. 2018;645:1334–1343.
- Lane-Petter W. The provision and use of pathogen-free laboratory animals. Proc R Soc Med. 1962;55:253–263.
- Marchesi JR, Ravel J. The vocabulary of microbiome research: a proposal. Microbiome. 2015;3(1):31.
- Lederberg J, McCray AT. ’Ome sweet ’omics - a genealogical treasury of words. Sci. 2001;15:8.
- Whipps J, Lewis K, Cooke R. Mycoparasitism and plant disease control. In: Burge N, editor. Fungi Biol. Control Syst. Manchester: Manchester University Press; 1988. p. 187.
- Ursell LK, Metcalf JL, Parfrey LW, et al. Defining the human microbiome. Nutr Rev. 2012;70(Suppl 1):S38–S44.
- Prescott SL. History of medicine: origin of the term microbiome and why it matters. Hum Microbiome J. 2017;4:24–25.
- Handelsman J, Rondon MR, Brady SF, et al. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem Biol. 1998;5(10):5.
- Handelsman J. Metagenomics: application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev. 2004;68(4):669–685.
- National Research Council (US). The new science of metagenomics: revealing the secrets of our microbial planet. Washington DC: National Academies Press; 2007.
