ABSTRACT
Ganoderma is a cosmopolitan genus that includes a great diversity of species. Many of them have been historically described based only on morphological characteristics; however, due to their morphological plasticity, there is no complete understanding about their relationship and taxonomic status. Commonly applied names, particularly in the southern Neotropics, come from species of North Hemisphere distribution (e.g. G. lucidum, G. resinaceum and G. applanatum). The objective of the present work was to perform a survey of Ganoderma species thriving in Uruguay. We aimed to identify and characterize them through molecular, morphological and ecological analysis. The results confirm the presence of four reddish laccate species first registered for Uruguay (G. dorsale, G. platense, G. martinicense and G. mexicanum), and one non-laccate species (G. australe s.l.) composed of two clades. The species are morphologically differentiated mainly by its stipe, pilear surface, context, pores, basidiospores and cutis cells. Regarding the ecological data, the species present differences in substrate preferences. In addition, a taxonomic discussion regarding phylogenetic relationships and taxonomic status of Uruguayan Ganoderma species is presented.
Introduction
Ganoderma (P. Karst.) harbors at least 220 species, being the most diverse genus of Ganodermataceae [Citation1–4]. However, due to their high diversity and phenotypic plasticity, the phylogenetic relationship and taxonomic status of many species remain unclear until now [Citation5,Citation6]. In the last 20 years, the use of molecular tools, mainly through the amplification and sequencing of Internal Transcribed Spacer (ITS), has been incorporated into systematic studies and into circumscription of Ganoderma species around the world. In this sense, through phylogenies based on molecular characters, the tendency in recent years has been to reinterpret the variations in characters of morphologically defined species, reinterpret the ecological relationships (relationship with hosts), determine their distribution and arrive at an understanding of the biogeographic processes that shape it [Citation5,Citation7–11].
Ganoderma species are morphologically characterized by the formation of sessile to stipitate basidiomata, with a glossy reddish laccate to opaque non-laccate cover, ellipsoid to ovoid double-walled basidiospores with truncated apex and endosporium with columnar ornamentations [Citation12]. This cosmopolitan genus is comprised of parasitic and saprophytic species that decay the wood of plants from temperate and tropical areas around the world [Citation6,Citation13,Citation14]. These fungi are described as white rot decayers that play a critical role in the dynamics of wood decomposition in tropical forests [Citation15]. Moreover, they are the main cause of tree deterioration in public ornamental and commercial plantations [Citation10,Citation15–18].
In the last decades, some researchers have tried to elucidate the diversity of Ganoderma genus in the Neotropics, particularly Bazzalo and Wright [Citation19], Gilbertson and Ryvarden [Citation20], Gottlieb and Wright [Citation21,Citation22], Gottlieb et al. [Citation7], Ryvarden [Citation12,Citation23], Torres-Torres and Dávalos [Citation24], Torres-Torres et al. [Citation25,Citation26], and more recently Cabarroi-Hernández et al. [Citation6] and Loyd et al. [Citation11] for the northern limit of Neotropical distribution. Uruguay in particular harbors a great biodiversity due to its transitional condition and ecoregions diversity [Citation27]. Some Ganoderma species have been historically recorded in Uruguay (), while Gazzano [Citation28,Citation29] and Martinez [Citation30] have made more recent contributions to the diversity of the genus in Uruguay.
Table 1. Ganoderma species recorded in Uruguay
Some Uruguayan cited species as G. applanatum (Pers.) Pat. (= G. lipsiense (Batsch) G.F. Atk.), G. sessile Murrill, G. lucidum P. Karst. and G. resinaceum Boud were distributed out of the southern Neotropics [Citation14,Citation35,Citation36], while others as G. lorenzianum (Kalchbr.) Pat., G. nitens (Fr.) Pat. lack of phylogenetic studies and their taxonomic status is unclear. Until now, there is no complete understanding on the systematic of the Ganoderma genus in Uruguay and its species diversity remains unknown.
The objective of the present work was to perform an updated survey of the Ganoderma species thriving in Uruguay and characterize them through molecular, morphological and ecological analysis. We also aimed to discuss the taxonomic status of the species of Uruguay and contrast with previous reports. We hypothesized that the Ganoderma specimens of Uruguay correspond to native species that have not been previously reported for the country. Those species are different from those registered to date (with names of species described from the Northern Hemisphere).
Materials and methods
Fungal specimens, basidiomata description and host characterization
Fresh Ganoderma basidiomata were collected from indigenous and urban ecosystems of Uruguay, during 2017 and 2018 field expeditions. Geographic location, host species and substrate condition (living tree, dead trunk, roots and stump) data were taken at the collection site following Urcelay and Robledo [Citation37]. For fresh basidiomata, small pieces were aseptically taken from the context, placed into 2% malt extract agar (MEA) and incubated in darkness at 25°C. Pure cultures and basidiomata were deposited in MVHC. In addition, specimens from national herbaria (MVHC and MVM) were examined. Herbarium acronyms follow Thiers [Citation38] (continuously updated, http://sweetgum.nybg.org/).
For morphological analyses and basidiomata identification, macroscopic and microscopic observations were made on basidiomata following the terminology and methodology according to authors [Citation6,Citation7,Citation19,Citation22–26]. Macroscopic features of basidiomata were analyzed and measured, particularly: pileus dimension, colour, texture, shape and appearance of surface, margin and stipe. The colour, presence of melanoid deposits and texture of context were also inspected and pores were described and measured (pores/mm). Microscopic features (basidiospores, chlamydospores in context, generative and somatic hyphae and cuticular cells) were analyzed with an optical microscope by mounting small sections of basidiomata with 5% KOH or Melzer’s Reagent (to test for dextrinoid or amyloid reaction). In particular, for the analysis of the hyphal system, sections of basidiomata were treated for 24–48 h in 3% NaOH at 50–60°C [Citation39]. Thirty basidiospores were measured from each specimen (length and width) and values were expressed as rank of mean values. Width was measured in the widest part of the spore and the length considered from the base to the truncated apex of the basidiospore. Then, Q ratio was calculated as the relation: length/width. Measures of cuticular cells were made from the middle part of the basidiomes.
Host relationships of each Ganoderma species in Uruguay were characterized by host range and native/exotic status. The preference of substrate condition was analyzed through the relative frequency of each Ganoderma species in each substrate condition (LT = stem of living tree, DT = dead trunks, S = stumps and R = soil, arising from roots of living or dead trees) following Urcelay and Robledo [Citation37]. Then, substrate preference was determined by transforming this relative frequency into a percentage.
DNA extraction, PCR and sequencing
DNA extractions from pure cultures were performed, using the CTAB protocol of Doyle and Doyle [Citation40] with modifications [Citation41]. The PCR reaction of the ITS region (including ITS1, 5.8s and ITS2) was performed. The following PCR primers were alternatively used: ITS1/ITS4 [Citation42], ITS1-F/ITS4-B [Citation43], and ITS4/ITS5 [Citation42]. The PCR mixture was prepared in a 25 μl final volume, with 2 μl of genomic DNA solution (10 ng), 16 μl of mQ water, 0.25 μl of Taq polymerase (1 U), 0.5 μl of each primer (10 mM), 0.7 μl of 50 mM MgCl2, 2.5 μl of dntps (2.5 mM) and 2.5 μl (10X) of buffer. PCR reactions were performed in a MultiGene Optimax thermocycler (Labnet International Inc) with the cycling conditions as follows: 3 min at 94°C, followed by 35 cycles, each consisting of 60 s at 94°C, 45 s at 50°C, 60 s at 72°C, and a final extension step at 72°C for 5 min. PCR products were verified by electrophoresis in 1.0% agarose gels in TBE buffer, stained with EZ vision®One (Amresco®) and visualized under UV light transillumination. GeneRuler DNA Ladder Mix marker (Thermo) was used as molecular size marker. PCR products were purified and sequenced by Macrogen (Seoul, Korea). Sequences were submitted to GenBank ().
Table 2. Ganoderma species, specimens, location, Gen Bank accession numbers for ITS sequences and reference source
Phylogenetic analyses
The sequences obtained were manually edited (visual inspection of sequences and chromatograms, resolution of conflicts and pair the extremes) with Bioedit V.7.0.5.3 [Citation47] and incorporated into alignments with sequences of specimens from other parts of the world obtained from GenBank (). Multiple alignment was made using ProbCons 1.12 from the CIPRES Science Gateway [Citation48]. Subsequently, the best evolutionary model for each region (ITS1, 5.8S and ITS2) was estimated using the Corrected Akaike Informational Criteria (AICc), implemented by the jModelTest2 v.1.6 software [Citation49]. The phylogenetic analysis was conducted in two independent ways: Bayesian Inference (BI) and Maximum Likelihood (ML), performed with MrBayes 3.2.7 [Citation50] and RAxML 8.2.12 [Citation51], respectively, in CIPRES Science Gateway [Citation52]. Cristataspora coffeata (Murrill) Robledo, Costa-Rezende and de Madrignac Bonzi (FLOR 50933) and Foraminispora rugosa (Berk.) Costa-Rezende, Drechsler-Santos and Robledo (FLOR 52191 and HUEFS DHCR560) were used as outgroup [Citation4,Citation53]. For the BI, two independent runs were performed, starting with random trees, with four independent and simultaneous chains, 10,000,000 MCMC generations, and maintaining 1 tree every 1000 generations. Burn in discarded values was indicated as 0.25. The estimated models for each partition were incorporated, as indicated below (see Results). The average standard deviation of split frequencies was limited to below 0.01. Convergence of the Markov chains to a stationary distribution was visually inspected using the Tracer v.1. 7. 1 program [Citation54]. A GTRGAMMA nucleotide model and 1000 bootstrap iterations were indicated for the ML analysis. The rest of the parameter values were set by default. Since the topologies of trees obtained in each analysis were convergent, only the consensus BI tree is shown with values of Bayesian posterior probability (BPP) and ML bootstrap (ML) separated by cross bars (BPP/BS). A clade was considered strongly supported if it showed a 0.95 BPP and/or 80% BS [Citation55].
Results
Fungal specimens, basidiomata description and host characterization
A total of 163 Ganoderma specimens from collections generated in this work (n = 90), MVHC (n = 57) and MVM (n = 16) were morphologically and ecologically analyzed (). Ganoderma australe was the most commonly collected species, with 101 specimens. The 62 remaining specimens belong to four reddish laccate species: G. mexicanum (n = 3), G. martinicense (n = 10), G. platense (n = 22) and G. dorsale (n = 27).
Table 3. Morphological assessment of Ganoderma species found in Uruguay
Table 4. Number of specimens, distribution and host characterization (species, native/exotic status and substrate preferences) of Ganoderma species collected and studied in this work
Morphologically assigned species are presented in . Ganoderma species of Uruguay were primarily differentiated by their pilear surface into two groups: reddish laccate and non-laccate ( and ).
Figure 1. Basidiomata of Ganoderma species from Uruguay. G. martinicense: (a) (MVHC 5583) and (b) (MVHC 5635), G. platense: (c) (MVHC 5586) and (d) (MVHC 5687), G. mexicanum (MVHC 5652): (e) and (f), G. dorsale: (g) (MVHC 5648) and (h) (MVHC 5655), G. australe M2 (MVHC 5587): (i) G. australe M1 (MVHC 5582): (j).

Figure 2. Microscopic features of laccate Ganoderma species from Uruguay. Pilear cells: G. martinicense (MVHC 5635): (a,b), G. platense (MVHC 5687): (d–f), G. mexicanum (MVHC 5652): (g–h), G. dorsale (MVHC 5655): (j–l). Chlamydospores from fresh G. martinicense (MVHC 5635) culture in MEA: (c) and dextrinoid characteristic chlamydospores from the context of G. mexicanum (MVHC 5652): (i). Basidiospores from G. martinicense (MVHC 5635): (m), G. platense (MVHC 5687): (n), G. mexicanum (MVHC 5652): (o) and G. dorsale (MVHC 5655): (p).

The first group is composed of two species with great and robust basidiomes (G. martinicense and G. platense) and two species with smaller basidiomes (G. dorsale and G. mexicanum).
Ganoderma martinicense is characterized by a large, commonly substipitate basidiomata with a tuberculous concentric zonated pilear surface and conspicuous melanoid deposits in the homogeneous context. Microscopically, it is characterized by the presence of a distinct smooth basidiospores ornamentation, formed by free pillars and cutis cells with weak reaction and spheroid-pedunculated shape. On the other hand, G. platense basidiomata are smaller, semi-orbicular zonate, homogeneous colored and always sessile. Its context is also different, presenting extremely inconspicuous melanoid lines and microscopically characterized by particular cutis elements with apical constrictions ().
Ganoderma mexicanum produces stylized, flabelliform, laterally and horizontally stipitate basidiomata characterized by an almost light-colored context and basidiospores with fine ornamentation, whereas G. dorsale produces stylized, spatuliform, shell-shaped and almost laterally and vertically stipitate basidiomata with no homogeneous distinctive context. Its basidiospores present notorious rough ornamentation and almost cylindrical cutis cells with a strong amyloid reaction ().
Specimens with non-laccate surfaces are morphologically very similar, discernible however by their pilear surface, which is distinctly tubercular in G. australe M1 and distinctly zonate in Ganoderma australe M2.
Herbaria specimens showed a morphology consistent with the species recorded in this study. In that sense, G. lucidum is a name previously used for specimens corresponding to the G. dorsale, G. martinicense, G. platense and G. mexicanum species. The name G. resinaceum was previously used to name specimens corresponding to G. platense and G. martinicense. The names G. lipsiense, G. applanatum and G. marmoratum were used to refer to specimens of G. australe in the broad sense (from now on sensu lato or s.l.). A morphological key for Ganoderma species found in Uruguay is presented below.
Host relationships of each Ganoderma species in Uruguay and presence in distinct departments are presented in . Ganoderma martinicense and G. australe were found alternatively in several departments growing on native or exotic trees. On the other hand, G. platense was only recorded in the southeast (Montevideo, Canelones and Maldonado departments), growing preferentially on exotic trees, G. mexicanum was only found on dead stems of native forest, and G. dorsale, almost exclusively on native trees. Regarding their substrate preferences, G. mexicanum and G. australe M2 were found preferentially on dead stem wood (100% and 46%, respectively); G. martinicense and G. dorsale, preferentially on roots (87% and 58%, respectively); and G. australe M1 and G. platense, preferentially on live stems (45% and 85%, respectively).
Phylogenetic analyses
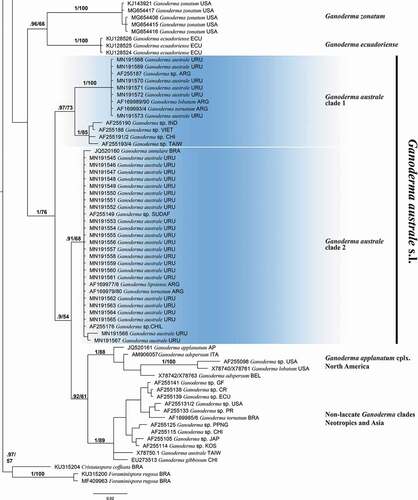
A total of 53 new sequences were generated from Uruguayan specimens. The dataset alignment resulted in 140 DNA sequences comprising 639 bp. The best evolutionary models for each partition were as follows: K80 + G (ITS1), JC (5.8 S), K80 + G (ITS 2). The partition scheme was K80 + G (ITS1) with -lnL = 1373.0169, and equal base frequencies as follows: A = 0.25, C = 0.25, G = 0.25, T = 0.25, JC (5.8 S) with -lnL = 300.8947 with equal base frequencies, and K80 + G (ITS 2) with 1498.7406 and equal base frequencies. The Bayesian Inference consensus tree is presented in .
Figure 3. Consensus Bayesian Inference tree based on ITS sequence data used for positioning Ganoderma species found in Uruguay (highlighted in light blue). Values of Bayesian and Maximum likelihood are presented as PPB/BS. Cristataspora coffeata (FLOR 50933) and Foraminispora rugosa (FLOR 52191 and HUEFS DHCR560) were used as outgroup.
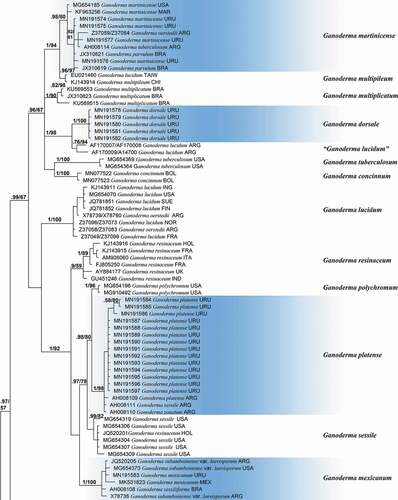
A total of 19 supported clades were recovered through a phylogenetic analyses (). Sequences corresponding to Uruguayan specimens were distributed in six terminal clades representing the following species (BPP/BS): Ganoderma platense Speg. (1/98), Ganoderma dorsale (Lloyd) Torrend (1/100), G. martinicense Welti & Courtec. (0.98/80), G. mexicanum Pat. (1/100) and Ganoderma australe s.l. composed of two terminal clades: one of them termed clade 1 (0.97/73), composed of specimens of G. australe M1 and the other one termed clade 2 (0.91/68) and composed of specimens of G. australe M2. The remaining clades represent next species: G. multipileum Hou. (0.96/97), G. multiplicatum (Mont.) Pat. (1/90), “G. lucidum” sensu authors [Citation22] (0.76/94), G. tuberculosum Murrill (1/100), G. concinnum Ryvarden (1/100), G. lucidum (1/100), G. resinaceum (0.9/59), G. polychromum Murrill (1/96), G. sessile (0.99/82), G. zonatum Murrill (1/100), G. ecuadoriense W.A. Salazar, C.W. Barnes & Ordoñez (1/100) and two unnamed taxa of the G. australe/applanatum complex from the Neotropic and Asia (1/89) and the Northern Hemisphere (1/88).
1. Reddish to wine-purplish pileus surface, glossy to opaque2
1´. Greyish brown pileus surface, matte, never reddishG. australe s.l. 5
2. Usually robust basidiomata, mostly sessile to occasionally substipitate3
2´. Stylized basidiomata, slender, laterally to eccentrically stipitate4
3. Pores 4–6/mm, basidiospores with thick endosporic pillarsG. martinicense
3´. Pores 3–4/mm, basidiospores with thin and tiny endosporic pillarsG. platense
4. Vertically stipitate basidiomata, brownish context and basidiospores with prominent endosporic pillarsG. dorsale
4´. Horizontally stipitate basidiomata, cream colored context and basidiospores with tiny endosporic pillar. G. mexicanum
5. Conspicuosly, tuberculous pileus surface, sometimes presenting concentric zones…. G. australe M1
5´. Concentrically zonated to slightly tubercular pileus surfaceG. australe M2
Key to Ganoderma species from Uruguay
Discussion and integrative taxonomy
The taxonomic status of Ganoderma species in Uruguay was evaluated through ITS-based phylogenetic analyses in combination with morphological and ecological data. The topology recovered in our phylogenetic analyses is congruent with previous works [Citation4–6]. Three main clades were recovered. One of them is composed of Ganoderma species with reddish laccate basidiomata, stipe and pilear surface, including traditional G. lucidum and G. resinaceum complexes (0.99/67). The second one is composed of almost-sessile Ganoderma species with non-laccate pilear surface, including G. australe and G. applanatum complexes (1/76). The third one is a small subclade of 2 reddish laccate species including G. zonatum and G. ecuadoriense (0.96/66). Studied Uruguayan Ganoderma specimens are distributed in six clades representing species whose taxonomic resolutions are discussed below.
Ganoderma martinicense is characterized by developing short stipitate basidiomata of large dimensions (up to 30 cm upper view), with the pilear surface concentrically zonate, with melanoid incrustations in the context, pores 5–6/mm, basidiospores measuring 10–12 × 5–7 µm and non-amyloid pear-shaped to shortly cylindrical cutis cells. The species is distributed from SW North America through the Caribbean and the Neotropical Atlantic Rain Forests, [Citation11,Citation56], reaching Uruguay and NE Argentina. Ten of the studied specimens fit very well with that morphological description. Moreover, they are grouped with G. martinicense specimens, including the type in a strongly supported clade (1/88). Other names were used to label sequences of specimens that are conspecific with sequences of this lineage (), i.e. G. tuberculosum [Citation7], G. oerstedii (Fr.) Murrill [Citation13] and G. parvulum Murrill [Citation57]. Ganoderma tuberculosum and G. parvulum form two different, distant and unrelated lineages [Citation6,Citation11]. Ganoderma oerstedii has been described as presenting a context lacking resinous lines, longer basidiospores with semi-rough columnar ornamentations: 12–15 × 8–10 µm [Citation12] and 9–14 × 6–9 µm [Citation19]. The pileipellis is formed by irregular, lobed and branched cells with up to seven short, wide protuberances [Citation25]. Sequences of specimens from the type locality (San Juan de Puerto Rico) need to be included to determine the taxonomic status and the phylogenetic relationships of G oerstedii. The closest relative of Ganoderma martinicense is G. multipileum (1/94), an Asian species with small pores: 6–8/mm [Citation9].
Five sequences of Uruguayan specimens are grouped together in a distinct, well-supported clade (1/100), unrelated to any available sequence. Specimens of this clade are characterized by a stylized basidiomata, laterally to centrally slender stipite, shiny reddish orange to violet colored radial and concentric zonate pilear surface, and melanoid bands in the context. Microscopically, the cutis is composed of clavate cells () and basidiospores with thick, often anastomosed endosporic ornamentations. The macro-morphological features suggest some Neotropical taxa including Ganoderma elegantum Ryvarden [Citation12], G. concinnum [Citation23] and G. dorsale [Citation58]. Ganoderma elegantum is characterized by pores 6–7/mm, branched cutis cells (with up to six branches) and basidiospores with thin endosporic ornamentations [Citation12]. Ganoderma concinnum, recorded in Colombia and Bolivia, is characterized by the formation of basidiomata with slender stipes of up to 20 cm long and it forms a different phylogenetic lineage [Citation12,Citation53]. Ganoderma dorsale (= G. lucidum var. dorsale in accordance with Torres-Torres et al. [Citation25]) was described from Río Grande Do Sul in southern Brazil [Citation59]. The macro and micro morphological characteristics of the specimens of this lineage agree with the description of the type specimen of G. dorsale [Citation22,Citation25] and fit well with the studied specimens: basidiomata shape and size, presence of melanoid lines in the context, basidiospores’ shape and size, chemical reactions and shapes of pileipellis cells. Thus, considering morphology and distribution, Ganoderma dorsale is the most suitable name for the new clade of Uruguayan specimens. The closest relative is a phylogenetic clade named “G. lucidum” from NW Argentinean Yungas [Citation7]. More sequences and morphological reassessments are necessary to evaluate the taxonomic status of the “G. lucidum” clade sensu Gottlieb et al. [Citation7].
Ganoderma multiplicatum, also occurring in the Neotropics, is morphologically differentiated by its cutis cells with numerous protuberances [Citation25,Citation57,Citation60,Citation61]. The phylogenetic relationship between G. multiplicatum, G. martinicense, G. multipileum, G. dorsale, “G. lucidum”, G. tuberculosum and G. concinnum remains unsolved.
Ganoderma platense is characterized by developing sessile basidiomata, usually imbricated, with a semi-orbicular zonate pilear surface context, usually with inconspicuous and discontinuous melanoid deposits, thin dissepiments and large pores (2–4/mm), pileipellis formed by cylindrical to slightly claviform elements, with apical constrictions and basidiospores measuring 9–13 × 5–8 µm, with thin, endosporic ornamentations [Citation7,Citation22,Citation62]. The morphological characteristics of the studied specimens constituting this clade ( and 2) agree with the description of G. platense and this name is hereinafter applied to the clade. Originally described from Buenos Aires (Argentina), it is currently known for growing on Platanus acerifolia trees of urban ecosystems of its type locality and on stumps of gallery forests of the Parana and Uruguay rivers [Citation7,Citation22,Citation62]. Within the G. platense clade, two sequences correspond to specimens labelled as G. sessile and G. zonatum [Citation7]; however, both species were phylogenetically circumscribed to distinct North American clades and are characterized by growing on hardwoods wood and monocots, respectively [Citation11]. Ganoderma platense is grouped in the so called “resinaceum clade”, together with G. resinaceum s.l. in Cabarroi-Hernández et al. [Citation6], G. polychromum and G. sessile sensu Loyd et al. [Citation11]. Ganoderma resinaceum encompasses North American, European and Asian populations, including more than one phylogenetic species. Nevertheless G. resinaceum sensu auctores from Eurasian descriptions present cutis cells with laterally diverticulate branches [Citation22,Citation25,Citation63,Citation64]. It contrasts with G. platense, which presents cylindrical to slightly clavate cells with apical constrictions. Ganoderma polychromum (Copel.) Murrill is distributed in North America, growing on hardwoods and characterized by a context without melanoid deposits, smaller pores (4–5 mm) and larger basidiospores measuring 10.8–13.2 × 6–7.5 µm [Citation11].
Ganoderma mexicanum is known for its occurrence in Brazil, Colombia, Costa Rica, Cuba, French Guiana, Mexico, Nicaragua, South–eastern USA (Florida) [Citation6], and now reported in Uruguay.
Morphologically, the studied specimen presents the diagnostic characters of light-colored context with variably abundant dextrinoid, smooth chlamydospores [Citation6].
The sequences of non-laccate Ganoderma specimens collected in Uruguay were distributed in two clades: one of them with Southern Hemisphere specimens and the other one with specimens from South America and Asia [Citation5]. Although the morphological analysis did not allow us to discriminate the specimens of each clade, the pilear surface generally appears tubercular and non-zonate in G. australe M1 and concentrically zonate G. australe M2. In addition, there seems to be ecological differences regarding host preferences: G. australe M1 specimens develop basidiomata on living hosts, whereas G. australe M2 on dead hosts. It was established that species of the G. australe/applanatum complexes could have a recent origin (not earlier than 30 Ma), with a distribution pattern explained by a large-scale, episodic colonization model and subsequent distance isolation [Citation5]. The recent origin, in addition to the remaining interfertility between specimens from both clades [Citation5], may be the plausible explanation of the crypticity and lack of clear morphological differences for the specimens of the two clades of G. australe. Many names were proposed for sequence of specimens grouped in each clade: G. tornatum (Pers.) Bres., G. lobatum (Schwein.) G.F. Atk., G. annulare (Lloyd) Boedijn and G. lipsiense [Citation7]. Ganoderma annulare and G. tornatum are largely considered synonyms of G. australe, and Ganoderma lipsiense was long considered a synonym of G. applanatum [Citation21,Citation25]. Ganoderma applanatum type locality is in the Northern Hemisphere and previous phylogenetic analyses suggested that it could be represented by a Northern Hemisphere clade [Citation5,Citation8]. Morphologically, G. applanatum lacks melanoid lines in the context [Citation25], whereas all Uruguayan specimens present melanoid elements in the context. From the morphological, ecological and distribution data, M1 and M2 specimens should remain as G. australe s.l.
In this context, the previous records of Ganoderma in Uruguay () should be questioned, and due to the absence of herbarium specimens, only speculations can be made in relation to published descriptions. Records of G. lorenzianum [Citation65,Citation66] have characteristics that resemble G. mexicanum due their morphological similarities regarding light colored context (picture in reference [Citation67]), stipe and ovoid, smooth basidiospores (9–10 × 6–7 µm). G. lorenzianum is an earlier name than G. mexicanum but the description and picture offered in bibliography [Citation67] and the low number of specimens of G. mexicanum analyzed in Uruguay is insufficient to assess the epitypification of these species. Focused studies on specimens corresponding to this species are urgently needed. In Uruguay, Ganoderma orbiforme (Fr.) Ryvarden (=G. fornicatum (Fr.) Pat.) was recorded by Felippone [Citation66], but it currently forms a different phylogenetic clade [Citation57] and no specimens or sequences corresponding to this species were recorded in the country. Ganoderma nitens was recorded by Patouillard [Citation65] characterized by warty basidiospores, 10 × 7 µm, the type is currently lost [Citation68], so this record could be attributed to G. dorsale, a single reddish laccate Ganoderma species of Uruguay with warty spores.
Conclusions
An integrated comprehensive approach was carried out using morphological, molecular and ecological evidence to assess the diversity of Ganoderma in Uruguay. A total of five species (forming six supported clades) were found through the analysis of 163 basidiomata and 53 sequences. Among those species, G. martinicense, G. mexicanum, G. platense and G. dorsale were confirmed and identified for the first time in Uruguay. Particularly, G. platense and G. dorsale were first recovered in the phylogenetic analyses. On the other hand, non-laccate specimens were distributed in two clades and so far considered as G. australe s.l.
Supplemental Material
Download MS Word (16.4 KB)Acknowledgments
The authors kindly acknowledge Anaclara Cabrera Varela for her technical support in improving figures editions. Curators of CORD, MVM and MVHC herbaria are acknowledged for the loan of collections for this study. Daniel Newman, (ORCID: 0000-0002-5400-3691) is kindly acknowledged for discussions, comments and proofreading the English version of the manuscript. The assistance of Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) and Universidad Nacional de Córdoba, both of which supported the facilities used in this project, is also acknowledged. Authorities that granted permits to collect in Uruguay are kindly acknowledged.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Karsten PA. Enumeralio boletinearum et polyporearum fennicarum, systemate novo dispositarum. Rev Mycol. 1881;3:16‒19.
- Murrill WA. Tomophagus for dendrophagus. Torreya. 1905;5:197.
- Moncalvo JM, Ryvarden L. A nomenclatural study of the Ganodermataceae. Oslo (Norway): Fungiflora; 1997.
- Costa-Rezende DH, Robledo GL, Góes-Neto A, et al. Morphological reassessment and molecular phylogenetic analyses of Amauroderma s.lat. raised new perspectives in the generic classification of the Ganodermataceae family. Persoonia. 2017;39:254‒269.
- Moncalvo JM, Buchanan PK. Molecular evidence for long distance dispersal across the Southern Hemisphere in the Ganoderma applanatum-australe species complex (Basidiomycota). Mycol Res. 2008;112(4):425‒436.
- Cabarroi-Hernández M, Villalobos-Arámbula AR, Torres-Torres MG, et al. The Ganoderma weberianum-resinaceum lineage: multilocus phylogenetic analysis and morphology confirm G. mexicanum and G. parvulum in the Neotropics. MycoKeys. 2019;59:95.
- Gottlieb AM, Ferrer E, Wright JE. rDNA analyses as an aid to the taxonomy of species of Ganoderma. Mycol Res. 2000;104(9):1033‒1045.
- Moncalvo JM. Systematics of Ganoderma. Ganoderma diseases of perennial crops; 2000.
- Wang DM, Wu SH, Su CH, et al. Ganoderma multipileum, the correct name for ‘G. lucidum’in tropical Asia. Bot Stud. 2009;50:451‒458.
- Coetzee M, Marincowitz S, Muthelo VG, et al. Ganoderma species, including new taxa associated with root rot of the iconic Jacaranda mimosifolia in Pretoria, South Africa. IMA Fungus. 2015;6(1):249‒256.
- Loyd AL, Barnes CW, Held BW, et al. Elucidating “lucidum”: distinguishing the diverse laccate Ganoderma species of the United States. PloS One. 2018;13(7):e0199738.
- Ryvarden L. Neotropical polypores: part 1: introduction, Ganodermataceae & Hymenochaetaceae. Oslo (Norway): Fungiflora; 2004.
- Moncalvo JM, Wang HF, Hseu RS. Gene phylogeny of the Ganoderma lucidum complex based on ribosomal DNA sequences. Comparison with traditional taxonomic characters. Mycol Res. 1995;99(12):1489‒1499.
- Cao Y, Wu SH, Dai YC. Species clarification of the prize medicinal Ganoderma mushroom “Lingzhi”. Fungal Divers. 2012;56(1):49‒62.
- Papp V. Global diversity of the Genus Ganoderma. In: Sridhar KR, and Deshmukh SK, editors. Advances in Macrofungi: diversity, ecology and biotechnology. 10–33. CRC Press; 2019 Jan 30.
- Glen M, Bougher NL, Francis AA, et al. Ganoderma and Amauroderma species associated with root-rot disease of Acacia mangium plantation trees in Indonesia and Malaysia. APP. 2009;38(4):345‒356.
- Rajchenberg M, Robledo G. Pathogenic polypores in Argentina. Forest Pathol. 2013;43(3):171‒184.
- Urcelay C, Robledo G, Heredia F, et al. Hongos de la madera en el arbolado urbano de Córdoba. Córdoba (Argentina): Instituto Multidisciplinario de Biología Vegetal (UNC-CONICET); 2012.
- Bazzalo ME, Wright JE. Survey of the Argentine species of the Ganoderma lucidum complex. Mycotaxon. 1982;16:295‒325.
- Gilbertson RL, Ryvarden L. North American polypores 1–2. Oslo (Norway): Fungiflora; 1986‒1987.
- Gottlieb AM, Wright JE. Taxonomy of Ganoderma from Southern South America: subgenus Elfvingia. Mycol Res. 1999;103:1289‒1298.
- Gottlieb AM, Wright JE. Taxonomy of Ganoderma from Southern South America: subgenus Ganoderma. Mycol Res. 1999;103:661‒673.
- Ryvarden L. Studies in neotropical polypores 2: a preliminary key to neotropical species of Ganoderma with a laccate pileus. Mycologia. 2000;92(1):180‒191.
- Torres-Torres MG, Guzmán-Dávalos L. The morphology of Ganoderma species with a laccate surface. Mycotaxon. 2012;119(1):201‒216.
- Torres-Torres MG, Guzmán-Dávalos L, de Mello Gugliotta A. Ganoderma in Brazil: known species and new records. Mycotaxon. 2013;121(1):93‒132.
- Torres-Torres MG, Ryvarden L, Guzmán-Dávalos L. Ganoderma subgénero Ganoderma en México. Rev Mex Micol. 2015;41:27‒45.
- Brazeiro A. Eco-regiones de Uruguay: biodiversidad, presiones y conservación: aportes a la Estrategia Nacional de Biodiversidad. Montevideo (Uruguay): Facultad de Ciencias, UDELAR; 2015.
- Gazzano S. Notas sobre Basidiomycetes xilófilos del Uruguay. X. Hongos Aphyllophorales de la Región E y NE (Departamentos de Cerro Largo, Rivera, y Treinta y Tres). Comun Bot Mus Hist Nat Montevideo. 2001;6(119):1‒10.
- Gazzano S. Notas sobre Basidiomycetes xilófilos del Uruguay. XIII. Aphyllophorales (Basidiomycota, Opistokonta) de la región Litoral Oesteeste y Noroeste de Uruguay. Comun Bot Mus Hist Nat Montevideo. 2010;138(7). 1–16.
- Martínez Kopp S. Comunidades de Basidiomycetes lignícolas en bosques nativos de Uruguay y factores que condicionan su composición. Montevideo (Uruguay): Universidad Nacional de Córdoba; 2014.
- Herter G. Florula Uruguayensis. Plantae Avasculares. Ostenia. Colección de Trabajos Botánicos dedicados a Don Cornelio Osten. 1933;7(84):1‒13.
- Gazzano S. Notas sobre Basidiomycetes xilófilos del Uruguay. VIII. Registro de Aphyllophorales y sus sustratos arbóreos. Comun Bot Mus Hist Nat Montevideo. 1998;6(109):1‒12.
- Spegazzini C. Fungi Argentini: novi v. critici. 1898;6.
- Wright JE, Bolontrade MF. An undesirable immigrant. Mycologist. 1994;8(1):14–15.
- Xing JH, Song J, Decock C, et al. Morphological characters and phylogenetic analysis reveal a new species within the Ganoderma lucidum complex from South Africa. Phytotaxa. 2016;266(2):115‒124.
- Richter CWK, Kirk PM, Stadler M. An assessment of the taxonomy and chemotaxonomy of Ganoderma. Fungal Diversity. 2015;71(1):1–15.
- Urcelay C, Robledo G. Community structure of polypores (Basidiomycota) in Andean alder wood in Argentina: functional groups among wood-decay fungi? Austral Ecol. 2004;29(4):471‒476.
- Thiers B. Index Herbariorum: a global directory of public herbaria and associated staff. New York Botanical Garden’s Virtual Herbarium; 2020 [cited 2020 Mar 31]. Available from: http://sweetgum.nybg.org/science/ih/
- Gómez-Montoya N, Rajchenberg M, Robledo GL. Aegis boa (Polyporales, Basidiomycota) a new neotropical genus and species based on morphological data and phylogenetic evidences. Mycosphere. 2017;8(6):1261‒1269.
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemistry. 1987;19:11‒15.
- Morera G, Robledo G, Ferreira-Lopes V, et al. South American Fomitiporia (Hymenochaetaceae, Basidiomycota) ‘jump on’ exotic living trees revealed by multi-gene phylogenetic analysis. Phytotaxa. 2017;321(3):277‒286.
- White TJ, Bruns T, Lee SJWT, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR protocols: a guide to methods and applications. New York (NY): Academic Press, Inc.; 1990. p. 315‒322.
- Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes‐application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2(2):113‒118.
- Guglielmo F, Gonthier P, Garbelotto M, et al. A PCR-based method for the identification of important wood rotting fungal taxa within Ganoderma, Inonotus sl and Phellinus sl. FEMS Microbiol Lett. 2008;282(2):228‒237.
- Crous PW, Wingfield MJ, Richardson DM, et al. Fungal Planet description sheets. Persoonia. 2016;36(316):400‒468.
- Zhou LW, Cao Y, Wu SH, et al. Global diversity of the Ganoderma lucidum complex (Ganodermataceae, Polyporales) inferred from morphology and multilocus phylogeny. Phytochemistry. 2015;114:7–15.
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In: Nucleic acids symposium series. Vol. 41; London: Information Retrieval Ltd., c1979-c2000; 1999. p. 95‒98.
- Do CB, Mahabhashyam MS, Brudno M, et al. ProbCons: probabilistic consistency-based multiple sequence alignment. Genome Res. 2005;15(2):330–340.
- Darriba D, Taboada GL, Doallo R, et al. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9(8):772.
- Ronquist F, Huelsenbeck JP. MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19(12):1572‒1574.
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688‒2690.
- Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: 2010 gateway computing environments workshop (GCE); IEEE; 2010 Nov 14. p. 1–8.
- Costa-Rezende DH, Robledo GL, Drechsler-Santos ER, et al. Taxonomy and phylogeny of polypores with ganodermatoid basidiospores (Ganodermataceae). Mycol Prog. 2020;19(8): 725 ‒41. DOI:https://doi.org/10.1007/s11557-020-01589-1
- Rambaut A, Drummond AJ, Xie D, et al. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst Biol. 2018;67(5):901.
- Hyde KD, Udayanga D, Manamgoda DS, et al. Incorporating molecular data in fungal systematics: a guide for aspiring researchers. 2013; arXiv preprint arXiv:1302.3244.
- Welti S, Courtecuisse R. The Ganodermataceae in the French West Indies (Guadeloupe and Martinique). Fungal Divers. 2010;43:103‒126.
- Correia de Lima Júnior N, Baptista Gibertoni T, Malosso E. Delimitation of some neotropical laccate Ganoderma (Ganodermataceae): molecular phylogeny and morphology. Rev Biol Trop. 2014;62(3):1197‒1208.
- Torrend C. Les Polyporacées du Brésil. I. Polyporacées stipités. Brotéria, Sér Bot. 1920;18:121‒143.
- Lloyd CG. Synopsis of the section Apus of the genus Polyporus. Lloyd Library; 1915. p. 4.
- Bolaños AC, Bononi VLR, Gugliotta de Mello A. New records of Ganoderma multiplicatum (Mont.) Pat. (Polyporales, Basidiomycota) from Colombia and its geographic distribution in South America. Check List. 2016;12(4). DOI:https://doi.org/10.15560/12.4.1948
- Steyaert RL. Study of some Ganoderma species. Bull Jard Bot Belg. 1980;135–186.
- Spegazzini C. Observations on and additions to Argentinian mycology. Bol Acad Nac Ci. 1926;28.
- Mohanty PS, Harsh NSK, Pandey A. First report of Ganoderma resinaceum and G. weberianum from north India based on ITS sequence analysis and micromorphology. Mycosphere. 2011;2(4):469–474.
- Ryvarden L, Gilbertson RL. European polypores: part 1: abortiporus-Lindtneria. Fungiflora A/S; 1993.
- Patouillard N. Le genre Ganoderma. Bull Soc Mycol France. 1889;5:64‒80.
- Felippone F. Contribution á la flore mycologique de l´Uruguay. Ann Cryptog Exot. 1928;1(4):338‒348.
- Kalchbrenner K. Szibériai és délamerikai gombák: fungi e Sibiria et America Australi [Értekezések a természettudományok köréből]. Budapest: Magyar Tudományos Akadémia Könyvkiadó Hivatala; 1878; 8. p. 16.
- Ryvarden L. Type studies in Polyporaceae 36, species described by E. In: Fries Synopsys Fungorum. Vol. 39; 2019.