ABSTRACT
Gaiadendron punctatum (Ruiz & Pav.) G. Don. exhibits a wide morphological variation and geographical distribution, ranging from Nicaragua to Bolivia. Reported polymorphism and the existence of more than 20 synonyms for Gaiadendron punctatum might indicate that there is either a cryptic complex, or that this species shows great variation. Populations from Northwestern Colombia were sampled to evaluate if local morphological variation was reflected in molecular variation. Two chloroplast regions were used, and haplotype networks were performed for each region. Also, reproductive and vegetative characters were compared between populations. Three different morphotypes were observed according to examined variables. DNA sequences showed some nucleotide substitutions and indels that characterized some of the morphotypes. Network analyses group together individuals of the same morphotype for both regions. We found evidence that the three different morphotypes here described showed some degree of genetic identity. Further work is needed for sampling along the whole distribution range of this species, to propose hypotheses about different entities or the existence of a single polymorphic taxon with altitudinal ecotypes.
Gaiadendron punctatum (Ruiz & Pav.) G. Don. exhibe una amplia variación morfológica y distribución geográfica, desde Nicaragua hasta Bolivia. El polimorfismo reportado y la existencia de más de 20 sinónimos para Gaiadendron punctatum podría indicar que existe un complejo críptico o que esta especie muestra una gran variación. Se muestrearon poblaciones del Noroccidente de Colombia para evaluar si la variación morfológica local se reflejaba en variación molecular. Se usaron dos regiones cloroplastídicas y se realizaron redes de haplotipos para cada región, también se compararon caracteres vegetativos y reproductivos entre poblaciones. Se observaron tres morfotipos diferentes de acuerdo con las variables examinadas. Las secuencias de ADN mostraron algunas sustituciones nucleotídicas, así como indels que caracterizaron algunos de los morfotipos. El análisis de redes agrupa individuos del mismo morfotipo para ambas regiones. Se encontró evidencia de que los tres morfotipos diferentes aquí descritos mostraron algún grado de identidad genética. Más trabajos se requieren, muestreando a través del rango completo de distribución de esta especie para proponer hipótesis sobre diferentes entidades o la existencia de un solo taxón polimórfico con ecotipos altitudinales.
Introduction
Gaiadendron punctatum (Ruiz & Pav.) G. Don. (Loranthaceae) was described from Perú in 1834 and it is distributed from Nicaragua to Bolivia [Citation1]. Polymorphisms of this species in vegetative and floral characters have been observed by Barlow and Wiens [Citation2], Kuijt and Lye [Citation3], Nickrent et al. [Citation4] and Roldán [Citation5]. Different corolla colors have been reported for this species by several authors, varying from yellow-orange [Citation2,Citation6–9], to white [Citation2,Citation6,Citation7,Citation9].
Several studies have evaluated morphological and molecular variation in parasitic plant species through different approaches, using both chloroplast and nuclear regions [Citation10–15]. Even, yellow-flowered populations of G. punctatum from Costa Rica were analyzed by Barlow and Wiens [Citation16] through a karyological study, in which authors observed intraspecific flower variation, where flowers growing in higher elevations were longer than flowers from lower elevations.
Since the publication of Gaiadendron punctatum in 1834, more than 20 names have been published and considered as synonyms of G. punctatum by Barlow and Wiens [Citation2] and Kuijt [Citation6,Citation17]. Gaiadendron G. Don was considered a monotypic genus until the publication of Gaiadendron coronatum Kuijt, in 2015. This new species occurs in Perú, and it can be recognized by the ellipsoid shape of the fruit with a persistent, prominent, yellow-greened tubular calyx and the presence of a crowned calyculus [Citation18]; the number of synonyms of Gaiadendron punctatum might indicate a diagnostic problem at morphological level; where either ecotypes of this species might be occurring, or this variation might indicate the existence of different taxa under a cryptic complex.
Field work in northwestern Colombia allowed the recognition of morphological variation among populations of Gaiadendron punctatum regarding leaf and flower characters. Considering the reported polymorphism of this species and the wide altitudinal range in which it occurs, we evaluated if local morphological variation was associated with genetic variation in some populations from Central and Western cordilleras of Colombian Andes, using two chloroplast regions.
Methods
Sampling
Sampling was made during 10 field explorations in different locations at northern, central and western cordilleras of Colombia. Close to 200 herbarium sheets of G. punctatum from Colombian herbaria COAH, COL, FAUC, HUA, JAUM and MEDEL were revised to characterize morphotypes. Three samples from yellow corolla morphotype, four samples from white corolla morphotype and one sample from pink-white corolla morphotype were collected, leaf tissue from each specimen was collected and kept in silica gel until DNA extraction, this material was also used for morphological analyses.
Morphological analysis
Variation in leaf size and shape, as well as differences in corolla color and size were observed in living specimens and in herbarium collections. Therefore, leaf length, leaf width, leaf length-width proportion and pre-anthesis corolla length measurements were made for more than 90 herbarium specimens; leaf shape was assigned according to length–width proportion, as proposed in Hickey [Citation19].
An analysis of variance (one-way ANOVA) was performed to identify if significant variation existed in leaf and flower measurements between morphotypes. Normality assumptions of data were tested through the Shapiro–Wilk test and the data matrix was normalized through a logarithmic transformation. A Tukey HSD test was performed to evaluate differences between pairs of morphotypes and a PCA analysis was also made to summarize variation in morphological traits, where variables were scaled prior to the analysis. All statistical analyses were made in software R [Citation20] with packages Factoextra [Citation21], FactomineR [Citation22], ggpubr [Citation23] and stats [Citation20].
DNA extraction and sequencing
DNA extraction was performed with a CTAB protocol, proposed by Ivanova et al. [Citation24], using 25 mg of leaf tissue for each sample. PCRs were made to amplify two chloroplast regions (trnL and trnH-PsbA), using the primers described in Shaw et al. [Citation25] for trnH-psbA and in Taberlet et al. [Citation26] for the intron in trnL. PCR was carried out with the following conditions:
TrnL intron: 95°C 10 min [35 cycles: 95°C 30s, 50°C 30s, 72°C 2 min]
TrnH-PsbA: 80°C 5 min; [35 cycles: 94°C 30s, 56°C 40s, 72°C 1 min]; 72°C 10 min
PCR mix included: 1x Buffer, 2 mM MgCl2, 0.2 µM for each primer, 0.2 µM dNTP, 0.2 µL of BSA 0.4 µg/µL, 0.2 µL of Thermo Scientific Taq DNA Polymerase 5 U/µL, 2 µL of genomic DNA and water to a final volume of 20 µL.
PCR products were purified using the ExoSAP protocol from Duminil [Citation27] with Thermo Scientific FastAP Buffer and Exonuclease I. Amplified fragments were sequenced in both directions at Macrogen Inc. (Korea). DNA fragments were assembled and visually checked with software Geneious [Citation28]. All sequences generated in this study were deposited in GenBank (see Supplemental Material 1).
Haplotype networks
Median-joining networks were performed with regions trnH-PsbA and trnL intron, using software PopArt [Citation29], with eight sequences for trnH-psbA and ten sequences for trnL intron, two trnL sequences were downloaded from Genbank (see Supplemental Material 1).
Results
Morphological patterns in Gaiadendron punctatum
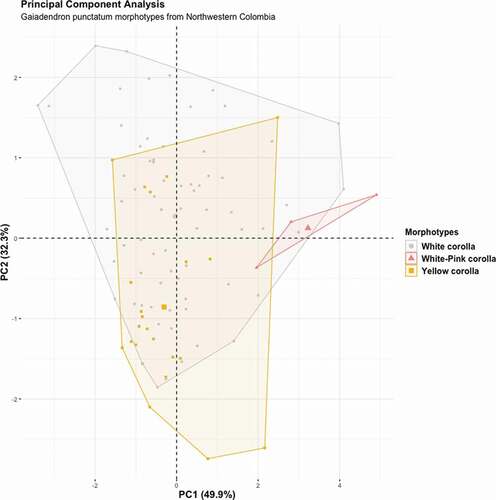
Three morphotypes of Gaiadendron punctatum were recognized based on corolla color and shape, in populations located at northern, central and western Andean cordilleras in Colombia (; ).
Table 1. Comparative morphological differences between the three Gaiadendron punctatum morphotypes of the northwestern Andes of Colombia
Figure 1. Occurrence of Gaiadendron punctatum sampled populations for haplotype analysis from northwestern Colombia
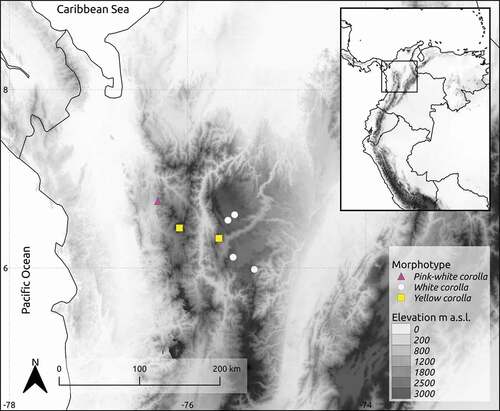
Figure 2. Morphological traits observed in three morphotypes of Gaiadendron punctatum. (a) White flowers; (b) White-pink flowers, (c) Yellow flowers, (d) Irregular calyculus in white flowers, (e) Irregular calyculus in white-pink flowers, (f) Membranous calyculus in yellow flowers. (g) Shoot and leaves of white-flowered morphotype, (h) Dots in leaf underside in white-pink flower morphotype, (i) Shoot and leaves of the yellow-flower morphotype. Calyculus is indicated with red arrows

White-Flowered Morphotype (, )
This morphotype is characterized by having tree habit, reaching 2–15 m of height. Leaves are elliptic (varying between widely elliptic and narrowly elliptic) to obovate (varying between narrowly obovate to oblanceolate), 2–12 cm length and 0.4–5.5 cm width. Flowers have a white-creamy corolla. Flower bud length ranges from 1–2.1 cm. The calyculus is a membrane with or without defined lobes or showing several teeth, this variation of calyculus was observed in flowers from the same inflorescence. This morphotype occurs at elevations ranging from 1100 to 3400 m a.s.l., in central and western cordilleras.
Yellow-Flowered Morphotype (, )
It includes trees or shrubs of 1.5–2 m of height, with elliptic leaves (being widely, narrowly or very narrowly elliptic), narrowly obovate to oblanceolate of 3.5–11 cm length and 1.2–4.4 cm width. The corolla color in this morphotype varies from yellow to orange, reaching a length from 1–2.6 cm. The calyculus usually has a membranous texture and it is also caducous, being sometimes persistent in the flower. This membranous calyculus was observed only in yellow-flowered plants. Populations occur at elevations between 2100 and 3400 m a.s.l. in both sampled cordilleras.
White-Pink Flowered Morphotype (, )
These plants are trees from 5–7 m height, with leaves between 7.4–13.2 cm length and 3.5–6.5 cm width. Leaf shape is elliptic. The petals in this morphotype are white, externally and pink-colored internally in the apex, with a flower bud length from 1–1.5 cm. The corolla develops a widening, which is not observed in yellow or white-creamy flowers. The calyculus is small, membranous and irregularly lobulate, similar to the White-Flowered Morphotype. Specimens were found in central and western cordilleras, between 1800 and 1950 m a.s.l.
Statistical analysis
Results of ANOVA tests showed that characters analyzed here, such as leaf length, leaf width, leaf length-width proportion and corolla length were significantly different (p < 0.05) between morphotypes (). The Tukey multiple comparison test evidenced that different characters are relevant to distinguish between morphotypes: leaf length and leaf length-width proportion distinguish between white and yellow corolla morphotypes, while leaf length and width are significantly different between white-pink and yellow corolla morphotypes, as well as white-pink and white corolla morphotypes. The white and yellow corolla morphotypes also differ significantly in corolla length.
Table 2. Results of PCA analysis, ANOVA and Tukey HSD test for leaf and flower characters of Gaiadendron punctatum morphotypes from Northwestern Colombia
PCA analysis shows that the two first components explain 82.2% of the total variation (PC1: 49.9%, PC2: 32.3%). PC1 mainly summarizes the variation in leaf length and leaf width and PC2 mainly summarizes the variation in length-width leaf proportion (which estimates leaf shape) and corolla length. The PCA plot of PC1 vs. PC2 does not show an evident separation of morphotypes in the space (). However, the white-pink corolla morphotype tends to differentiate in higher values for PC1 (leaf size) and the yellow corolla morphotype in lower values of PC2 (corolla length and leaf shape).
Nucleotide sequence diversity in chloroplast regions
Total number of sequences generated in this study were: eight sequences for White-Flowered Morphotype (four for trnH-psbA and four for trnL), six sequences for Yellow-Flowered Morphotype (three for trnH-psbA and three for trnL) and two sequences for White-Pink Flowered Morphotype (one for trnH-psbA and one for trnL).
Sequences from trnH-psbA region ranged between 455 and 558 bp in length and sequences from trnL intron region ranged between 299 and 334 bp in length. Variation between morphotypes was mostly observed in the trnH-psbA region.
The alignment obtained with the trnL region displayed three nucleotide differences between white corolla and yellow corolla sequences, at positions 55, 162 and 252; these same differences were found between yellow corolla sequences and the white-pink corolla sequence. No variation was observed between white corolla sequences and the white-pink sequence for this region ().
Table 3. Alignment of non-coding chloroplast region trnL intron indicating substitutions and indels in sequences of Gaiadendron punctatum morphotypes. Dashes represent gaps in position ranges or in individual positions; dots indicate the same nucleotides as for White Corolla 1
Six sites in the trnH-psbA matrix were found different between white corolla individuals and yellow corolla individuals at positions 67, 107–113, 119–124, 130–136, 231–237 and 285; the white-pink corolla sequence has four differences with the white corolla sequences in positions 107–113, 119–124, 130–136 and 285 ().
Table 4. Alignment of non-coding chloroplast region trnH-psbA indicating substitutions and indels in sequences of Gaiadendron punctatum morphotypes. Dashes represent gaps in position ranges or in individual positions; dots indicate the same nucleotides as for White Corolla 1
Two differences are observed between yellow corolla and white-pink corolla sequence in positions 67 and 231–237. In addition, the trnH-psbA white-pink corolla sequence has an indel at positions 316–324, and unique substitutions at positions 353–354 and 388 ().
Population analysis
Haplotype networks are shown in , based on chloroplast regions trnH-psbA and trnL intron, both regions are differently informative.
Figure 4. Median Joining haplotype networks of Gaiadendron punctatum populations for (a) trnL intron and (b) trnH-psbA. Lines represent mutational differences between haplotypes. Star indicates White-Pink Morphotype
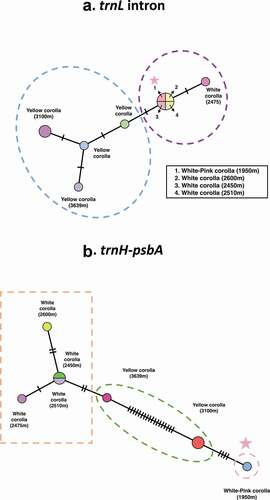
The haplotype network obtained with the trnL intron shows less variation among morphotypes. Yellow corolla sequences show more affinity among each other and form a clade ()); white-pink and white corolla sequences are grouped together. Less mutations between samples are seen in this network.
Sequences of trnH-psbA obtained for white corolla morphotype, display more affinity with each other than with yellow corolla sequences or with the white-pink corolla sequence ()). The yellow corolla sequences are central in the network.
Discussion
Some of the most contrasting features observed across local morphological variation found in G. punctatum are related to flower (color, shape of corolla and shape of calyculus) and leaf (size, shape and thickness) attributes. Here, we report a new variation for the species in corolla color, observing white flowers with pink petals in the apex and a widening in the base of the corolla.
Statistical analyses performed here support that several morphotypes of Gaiadendron punctatum might be observed locally. Leaf and flower measurements could be useful for analyzing variation in this species. Also, revising more herbarium specimens and collecting more individuals in the field, as well as evaluating further vegetative and reproductive characters could help to distinguish between these and other variations found in different regions, along the geographic distribution of this species.
Populations of G. punctatum with yellow flowers outside Colombia have been reported occurring in Costa Rica [Citation8,Citation30] and Ecuador [Citation6]. On the other hand, populations with white flowers are also found in Ecuador, but these are less common [Citation6], while white flowered populations are more widespread in northwestern Colombia and more frequently collected than yellow flowered or white-pink flowered populations.
trnH-psbA chloroplast region displayed more nucleotide changes in the alignment and it was more informative for the haplotype network. This region proved to be useful in this study, it is proposed as a universal DNA barcode in plants and it is recommended for phylogenetic analyses and species identification [Citation31–33], although it is short length and sometimes not informative enough [Citation34]. Therefore, using different chloroplast regions or nrDNA as ITS might add molecular information for this species.
Morphological variation found in Gaiadendron punctatum, indicates that local populations exhibit habit and corolla color differences, which are also separated by elevational gradient; it is noteworthy that white corolla populations are the most widespread throughout Colombian territory, they also occur in a wider altitudinal range. Yellow corolla populations occur at more restricted elevations (above 2000 m), and white-pink populations with few individuals collected, have been only registered in two locations from similar elevations. Rarity of this morphotype might be due to highly anthropic activity observed in the area where these populations occur.
Elevation trends in morphological characters as corolla length observed in this study () are consistent with those reported by Barlow and Wiens [Citation16], their results suggest that variation in corolla length corresponds to elevation ranges, instead of geographical patterns. Similarly, intraspecific variation in Colombian species of genus Anthurium (Araceae) has been studied, showing morphological differences in flower traits along an elevational gradient [Citation35].
Molecular variation found in the sampled populations of G. punctatum might indicate several phenomena occurring in this species, such as the existence of ecotypes following “Ecotypic divergence” concept, used in Armbruster [Citation36], cryptic species [Citation37] or a single polymorphic taxon. Haplotype networks obtained here show some divergence between these populations, as the nucleotide alignment reflects similarities between white flowered sequences, different from yellow-flowered sequences and even some unique characters in the only pink-flowered sequence included.
Studies published, as for example Deroo et al. [Citation38], show that few nucleotide substitutions in alignments from a single region, have been useful to differentiate species of Gallium (Rubiaceae). Nevertheless, a wider sampling of G. punctatum populations and molecular analyses including a higher number of individuals, might help to propose hypotheses about the molecular diversity in this species. Also, these studies would be necessary because of reported differences in chromosome numbers in G. punctatum [Citation16].
Three Gaiadendron punctatum morphotypes are described from northwestern Colombia, displaying local morphological variation, which was also observed in nucleotide variation, using two chloroplast regions. Further study using additional nuclear or chloroplast regions and including samples along the whole distribution range of this species, will allow a wider perspective to study the morphological variation in this taxon. Additional studies such as palynological, anatomical or phylogenetic, might help to characterize the overall variation found in G. punctatum, in order to propose different taxa under the name of G. punctatum or confirm if either this is a polymorphic taxon or different ecotypes are being observed.
Author contributions
F. Alzate-Guarín, I. Carmona-Gallego, and R. Vidal-Russell designed the study, I. Carmona-Gallego and F. Alzate-Guarín collected data and generated the sequences; I. Carmona-Gallego, F. Alzate-Guarín and R. Vidal-Russell analyzed the data and wrote the manuscript.
Supplemental Material
Download MS Excel (9.3 KB)Acknowledgments
We thank Francisco Javier Roldán, Mailyn González, Herber Sarrazola and Jhon Steven Murillo for their taxonomic help and suggestions. We also thank the staff at herbaria COAH, COL, FAUC, HUA, JAUM and MEDEL for granting access to study their Gaiadendron specimens. Authors thank two anonymous reviewers for their comments to improve previous versions of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Kuijt J, Hansen B. Loranthaceae. In: Kubitzki K, editor. The families and genera of vascular plants. New York: Springer International Publishing; 2015. p. 73–119.
- Barlow BA, Wiens D. The classification of the generic segregates of Phrygilanthus (= Notanthera) of the Loranthaceae. Brittonia. 1973;25(1):26–39.
- Kuijt J, Lye D. A preliminary survey of foliar sclerenchyma in neotropical Loranthaceae. Blumea. 2005;50(2):323–355.
- Nickrent DL, Malécot V, Vidal-Russell R, et al. A revised classification of Santalales. Taxon. 2010;59(2):538–558.
- Roldán FJ. Contribución al conocimiento de las lorantáceas del departamento de antioquia. [Contribution to the knowledge of Loranthaceae from the department of Antioquia]. Medellín: Universidad de Antioquia; 1993. Spanish.
- Kuijt J. Loranthaceae. In: Harling G, Sparre B, editors. Flora of Ecuador. Stockholm: Swedish Natural Science Research Council; 1986. p. 115–194.
- Kuijt J. Loranthaceae. In: Stevens WD, Ulloa-Ulloa C, Pool A, et al. editors. Flora de Nicaragua [Flora of Nicaragua]. St Louis: Missouri Botanical Garden Press; 2001. p. 1239–1246. Spanish.
- Morales JF. Loranthaceae. In: Hammel BE, Grayum M, Herrera C, et al. editors. Manual de plantas de Costa Rica [Manual of plants of Costa Rica]. St Louis: Missouri Botanical Garden Press; 2007. p. 218–235. Spanish.
- Alzate F, Idárraga A, Díaz O, et al. Flora de los bosques montanos de medellín. [Flora of montane forests of Medellín]. Medellín: Alcaldía de Medellín; 2013. Spanish.
- Amico GC, Nickrent DL. Population structure and phylogeography of the mistletoes Tristerix corymbosus and T. aphyllus (Loranthaceae) using chloroplast DNA sequence variation. Am J Bot. 2009;96(8):1571–1580.
- Laphitz RML, Ezcurra C, Vidal-Russell R. Cryptic species in the Andean hemiparasite Quinchamalium Chilense (Schoepfiaceae: Santalales). System Biodivers. 2018;16(3):260–270
- Vidal-Russell R. Phylogenetic relationships in Arjona (Schoepfiaceae), a hemiparasitic herb from Southern South America. Syst Bot. 2019;44(3):592–599.
- Amico GC, Vidal-Russell R, Garcia MA, et al. Evolutionary history of the South American mistletoe Tripodanthus (Loranthaceae) using nuclear and plastid markers. Syst Bot. 2012;37(1):218–225.
- Pérez‐Crespo MJ, Ornelas JF, González‐Rodríguez A, et al. Phylogeography and population differentiation in the Psittacanthus calyculatus (Loranthaceae) mistletoe: a complex scenario of climate–volcanism interaction along the Trans‐Mexican Volcanic Belt. J Biogeogr. 2017;44(11):2501–2514.
- Molvray M, Kores PJ, Chase MW. Phylogenetic relationships within Korthalsella (Viscaceae) based on nuclear ITS and plastid trnL‐F sequence data. Am J Bot. 1999;86(2):249–260.
- Barlow BA, Wiens D. The cytogeography of the loranthaceous mistletoes. Taxon. 1971;20(2–3):291–312.
- Kuijt J. Commentary on the mistletoes of Panama. Ann Mo Bot Gard. 1978;65(2):736–763.
- Kuijt J, Graham JG. Two new species of Loranthaceae from central Peru. Novon. 2015;24(2):173–178.
- Hickey LJ. Classification of the architecture of dicotyledonous leaves. Am J Bot. 1973;60(1):17–33.
- R Core Team. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing; 2020. Available from: https://www.R-project.org/
- Kassambara A, Mundt F. Factoextra: extract and visualize the results of multivariate data analyses. R package version 1.0.7; 2020. Available from: https://CRAN.R-project.org/package=factoextra
- Le S, Josse J, Husson F. FactoMineR: an R package for multivariate analysis. J Stat Softw. 2008;25(1):1–18.
- Kassambara A. ggpubr: ‘ggplot2ʹ based publication ready plots. R package version 0.4.0; 2020. Available from: https://CRAN.R-project.org/package=ggpubr
- Ivanova NV, Fazekas AJ, Hebert PD. Semi-automated, membrane-based protocol for DNA isolation from plants. Plant Mol Biol Rep. 2008;26(3):186–198.
- Shaw J, Lickey EB, Beck JT, et al. The tortoise and the hare ii: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am J Bot. 2005;92(1):142–166.
- Taberlet P, Coissac E, Pompanon F, et al. Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Res. 2007;35(3):e14–e14.
- Duminil J. Mitochondrial genome and plant taxonomy. In: Besse P, editor. Molecular plant taxonomy: methods and protocols. New York: Humana Press; 2014. p. 121–140.
- Kearse M, Moir R, Wilson A, et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(2):1647–1649.
- Leigh JW, Bryant D. POPART: full-feature software for haplotype network construction. Methods Ecol Evol. 2015;6(9):1110–1116
- Kuijt J. On the ecology and parasitism of the Costa Rican tree mistletoe, Gaiadendron punctatum (Ruiz & Pavon) G. Don. Can J Bot. 1963;41(6):927–938
- Kress WJ, Wurdack KJ, Zimmer EA, et al. Use of DNA barcodes to identify flowering plants. Proc Natl Acad Sci U S A. 2005;102(23):8369–8374.
- Pang X, Liu C, Shi L, et al. Utility of the trnH–psbA intergenic spacer region and its combinations as plant DNA barcodes: a meta-analysis. Plos One. 2012;7(11):e48833.
- Santos C, Pereira F. Identification of plant species using variable length chloroplast DNA sequences. Forensic Sci Int-Gen. 2018;36:1–12.
- Dong W, Liu J, Yu J, et al. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. Plos One. 2012;7(4):e35071.
- Cuartas-Hernández SE, Moreno-Betancur DJ, Gibernau M, et al. Contrasting patterns of floral size variation in two sympatric species of Anthurium along an elevation gradient in a tropical mountain forest. Int J Plant Sci. 2019;180(3):209–219.
- Armbruster WS. Floral specialization and angiosperm diversity: phenotypic divergence, fitness trade-offs and realized pollination accuracy. AoB Plants. 2014;6:1–24.
- Bickford D, Lohman DJ, Sodhi NS, et al. Cryptic species as a window on diversity and conservation. Trends Ecol Evol. 2007;22(3):148–155.
- Deroo AC, Eckstein P, Benaragama D, et al. Evaluation of Galium species and populations using morphological characters and molecular markers. Weed Res. 2018;59(1):28–38.