Abstract
We decoded the complete chloroplast DNA (cpDNA) sequence of the Wakame (Undaria pinnatifida), an important economic macroalga of the family Alariaceae, by using next-generation sequencing technology. The genome consists of 130 336 bp containing a pair of inverted repeats (IRs) of 4790 bp, which was separated by a large single-copy region and a small single-copy region of 77 821 and 42 934 bp, respectively. The genic regions account for 77.7% of whole cpDNA, and the GC content of the cpDNA was 30.6%. The U. pinnatifida cpDNA encodes 153 unigenes (129 protein-coding genes, 3 rRNA genes and 21 tRNA genes). There are 1 PCG (rpl33) and 1 tRNA genes (trnL) containing an intron. A phylogenetic analysis of the four complete cpDNA from Phaeophyceae showed that U. pinnatifida is closely related to Saccharina japonica with high bootstrap value supported. The complete cpDNA of U. pinnatifida provides essential and important DNA molecular data for further phylogenetic and evolutionary analysis for brown algae.
Introduction
Wakame (Undaria pinnatifida) is an important economic macroalga in East Asian countries (Yamanaka & Akiyama Citation1993). In China, its annual yield has been maintained around 500 000 tons in fresh weight in recent years, ranking second in brown seaweeds. Dalian, which is located in the southernmost of Liaodong peninsula, is the prime farming ground of this macroalga. Undaria pinnatifida is regarded as a healthy marine vegetable because it contains high content of nutrition (essential amino acid, vitamins and trace minerals) (Nisizawa et al. Citation1987; Taboada et al. Citation2013) and bioactive compounds (fucoidans) (Synytsya et al. Citation2010; Liu et al. Citation2012b). De novo transcriptome sequencing and assembly of the gametophyte of U. pinnatifida has been conducted and putative key genes involved in important biosynthetic pathway of fucoidan, alginate, mannitol, and laminarin were identified (Shan et al. Citation2015a). A high-density genetic linkage map has very recently been constructed and the sex linked locus was mapped for the first time in this macroalga (Shan et al. Citation2015b). In this study, we aimed to deduce the complete chloroplast genome to obtain essential sequence information for further research on genetics and evolution.
Sample of U. pinnatifida (voucher no. 474) was collected from Dalian, Liaoning province of China. Genomic DNA was extracted following the modified CTAB DNA extraction protocol (Attitalla Citation2011) and then subjected to build up genomic library and pair-end sequencing (2 × 300 bp) by MiSeq (Illumina, San Diego, CA). By using commercial software (Geneious V9, Auckland, New Zealand), about 3.9% (408,662 out of 10 472 286) raw reads were de novo assembled to produce circular form of complete cpDNA with about an average 974 × coverage.
Annotation of the assembled genome was performed with DOGMA (Wyman et al. Citation2004), cpGAVAS (Liu et al. Citation2012a) and manual inspected to predict protein-coding genes (PCGs), transfer RNA (tRNA) and ribosome RNA (rRNA) genes. The complete cpDNA of U. pinnatifida has a total length of 130 336 bp (GenBank KU200463) showing 93% identity to Saccharina japonica. It has a typical quadripartite structure including a pair of inverted repeats (IRa and IRb 4790 bp), separated by the small single-copy (SSC 42 934 bp) and large single-copy (LSC 77 821 bp) regions. The complete cpDNA of U. pinnatifida contains 153 unique genes consisting 21 transfer RNA, 3 ribosomal RNA, and 129 PCGs. Among 153 unique genes, there are two genes (rpl33 and trnL genes) are interrupted by one intron. The chloroplast genome consists 77.7% genic regions, and the overall GC content of the complete cpDNA is 30.6%. The GC content of IR regions is 44.8%, higher than LSC (29.6%) and SSC (29.3%) regions. The relative low GC content of LSC and SSC regions is due to low GC content in the PCGs and non-coding region.
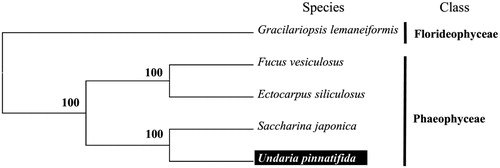
To validate the phylogenetic position of U. pinnatifida, we used MEGA6 (Tamura et al. Citation2013) software to construct a Maximum likelihood tree (with 500 bootstrap replicates) containing complete cpDNA of four algae in Phaeophyceae. Gracilariopsis lemaneiformis derived from Florideophyceae was used as outgroup for tree rooting. Result shows U. pinnatifida is closely related to Saccharina japonica with high bootstrap value supported (). In conclusion, the complete cpDNA of U. pinnatifida is decoded for the first time in this study and provides essential and important DNA molecular data for further phylogenetic and evolutionary analysis for Phaeophyceae.
Figure 1. Molecular phylogeny of Undaria pinnatifida and other related brown algae based on complete chloroplast genome. The complete chloroplast genome is downloaded from GenBank and the phylogenic tree is constructed by the maximum-likelihood method with 500 bootstrap replicates. The gene's accession number for tree construction is listed as follows: Gracilariopsis lemaneiformis (KU179794), Fucus vesiculosus (NC_016735), Ectocarpus siliculosus (NC_013498), Saccharina japonica (NC_018523) and Undaria pinnatifida (KU200463).
Disclosure statement
None of the authors report any conflict of interest. The authors alone are responsible for the content and writing of the paper.
Funding information
The research is supported from the grants from Natural Science Foundation of Zhejiang Province (LQ14B070002) and Zhejiang province science and Technology Department public technology research project of Agriculture (2015C32001).
References
- Attitalla IH. 2011. Modified CTAB method for high quality genomic DNA extraction from medicinal plants. Pakistan J Biol Sci. 14:998–999.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012a. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715.
- Liu F, Wang J, Chang AK, Liu B, Yang L, Li Q, Wang P, Zou X. 2012b. Fucoidan extract derived from Undaria pinnatifida inhibits angiogenesis by human umbilical vein endothelial cells. Phytomedicine. 19:797–803.
- Nisizawa K, Noda H, Kikuchi R, Watanabe T. 1987. The main seaweed foods in Japan. Paper presented at Twelfth International Seaweed Symposium, Springer.
- Shan T, Pang S, Li J, Li X. 2015a. De novo transcriptome analysis of the gametophyte of Undaria pinnatifida (Phaeophyceae). J Appl Phycol. 27:1011–1019.
- Shan T, Pang S, Li J, Li X, Su L. 2015b. Construction of a high-density genetic map and mapping of a sex-linked locus for the brown alga Undaria pinnatifida (Phaeophyceae) based on large scale marker development by specific length amplified fragment (SLAF) sequencing. BMC Genomics. 16:902.
- Synytsya A, Kim W-J, Kim S-M, Pohl R, Synytsya A, Kvasnička F, Čopíková J, Park YI. 2010. Structure and antitumour activity of fucoidan isolated from sporophyll of Korean brown seaweed Undaria pinnatifida. Carbohydrate Polym. 81:41–48.
- Taboada M, Millán R, Miguez M. 2013. Nutritional value of the marine algae wakame (Undaria pinnatifida) and nori (Porphyra purpurea) as food supplements. J Appl Phycol. 25:1271–1276.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yamanaka R, Akiyama K. 1993. Cultivation and utilization of Undaria pinnatifida (wakame) as food. J Appl Phycol. 5:249–253.
