Abstract
In this study, the complete mitogenome sequence of Galaxy Coral, Galaxea fascicularis (Cnidaria: Oculinidae), has been sequenced by next-generation sequencing method. The overall base composition of Galaxea fascicularis mitogenome is 24.9% for A, 14.0% for C, 24.3% for G and 36.8% for T and has low GC content of 38.3%. The assembled mitogenome, consisting of 18 751 bp, has unique 13 protein-coding genes (PCGs), three transfer RNAs and two ribosomal RNAs genes. The G. fascicularis mitogenome has the common mitogenome gene organization and feature of scleractinian coral. Among 13 PCGs, ND5 gene is interrupted by group I intron (12 022 bp). There are 12 genes embedded in ND5 group I intron (tRNA-Leu, ND1, CYTB, ND2, ND6, ATP6, ND4, 12S rRNA, COX3, COX2, ND4L and ND3). The complete mitogenome provides essential and important DNA molecular data for further phylogenetic and evolutionary analysis for stony coral.
The Galaxy Coral (Galaxea fascicularis) is common in a wide range of habitats and may be a dominant species on inshore fringing reefs. Small colonies are cushion shaped or low domes or are irregular. Large colonies, which frequently exceed five metres across, are columnar or massive. The colour is usually green, grey, red or brown, commonly with tentacles and septa of contrasting colours. Tentacles often have conspicuous white tips (Veron Citation2000). The establishment of G. fascicularis mitogenome is usefulness for further evolutionary and phylogenetic research of stony coral.
Samples (voucher no. 467) of G. fascicularis were collected from Hainan Island, China. The methods for genomic DNA extraction, library construction and next-generation sequencing were followed by our previous publication (Shen et al. Citation2014). Initially, the raw next-generation sequencing reads generated from MiSeq (Illumina, San Diego, CA). About 0.24% raw reads (7151 out of 2 986 720) were de novo assembly by using commercial software (Geneious V9, Auckland, New Zealand) to produce a single, circular form of complete mitogenome with about an average 113 X coverage.
The complete mitochondrial genome of G. fascicularis was 18 751 bp in size (GenBank: KU159433) and its overall base composition is 25.7% for A, 22.9% for C, 28.6% for G and 22.9% for T, showing 98% identities to the complete mitogenome of Euphyllia ancora (GenBank: JF825139). The complete mitogenome of G. fascicularis includes unique 13 protein-coding genes, three transfer RNA genes and two ribosomal RNA genes. The protein coding, rRNA and tRNA genes were predicted by using DOGMA (Wyman et al. Citation2004), ARWEN (Laslett & Canback Citation2008), MITOS (Bernt et al. Citation2013) and manual inspected. All genes, including 13 PCGs, tRNA and rRNA, were encoded on H-strand. All the 13 PCGs share the start codon ATG, except for COX3, ND3 and ND5 (with GTG start codon). All the 13 PCGs share the stop codon TAA, except for COX3, ATP6, ND3 and ND5 (with TAG stop codon). The longest one is ND5 gene (1836 bp) in all PCGs, whereas the shortest is ATP8 gene (219 bp). The size of small ribosomal RNA (12S rRNA) and large ribosomal RNA (16S rRNA) genes are 1186 bp and 2308 bp, respectively. Among 13 PCGs, ND5 gene is interrupted by group I intron (12 022 bp). There are 12 genes embedded in ND5 group I intron (tRNA-Leu, ND1, CYTB, ND2, ND6, ATP6, ND4, 12S rRNA, COX3, COX2, ND4L and ND3). Similar mitogenomic organization has been reported in previous studies in scleractinian coral (Medina et al. Citation2006; Fukami et al. Citation2007).
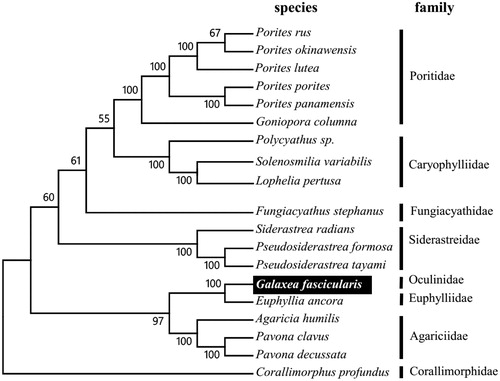
To validate the phylogenetic position of G. fascicularis, we used MEGA6 software (Tamura et al. Citation2013) to construct a maximum-likelihood tree (with 500 bootstrap replicates) containing complete mitogenomes of 18 species derived from seven different families in Scleractinia. Corallimorphus profundus derived from Corallimorphidae was used as out group for tree rooting. Result shows G. fascicularis showing close related to Hammer coral (Euphyllia ancora) with high bootstrap value supported (). In conclusion, the complete mitogenome of the G. fascicularis deduced in this study provides essential and important DNA molecular data for further phylogenetic and evolutionary analysis for stony coral phylogeny.
Figure 1. Molecular phylogeny of Galaxea fascicularis and other related species in Scleractinia based on complete mitogenome. The complete mitogenome is downloaded from GenBank and the phylogenic tree is constructed by maximum-likelihood method with 500 bootstrap replicates. The gene's accession number for tree construction is listed as follows: Porites rus (NC_027526), Porites okinawensis (NC_015644), Porites lutea (KU159432), Porites porites (NC_008166), Porites panamensis (NC_024182), Goniopora columna (NC_015643), Polycyathus sp. (NC_015642), Solenosmilia variabilis (NC_025472), Lophelia pertusa (NC_015143), Fungiacyathus stephanus (NC_015640), Siderastrea radians (NC_008167), Pseudosiderastrea formosa (NC_026530), Pseudosiderastrea tayami (NC_026531), Galaxea fascicularis (KU159433), Euphyllia ancora (NC_015641), Agaricia humilis (NC_008160), Pavona clavus (NC_008165), Pavona decussata (NC_026527) and Corallimorphus profundus (NC_027105)
Disclosure statement
The authors report no conflicts of interest.
Funding information
The authors alone are responsible for the content and writing of the paper. This research was supported by grants from the National Natural Science Foundation of China (41406161), the Basic Science Research Fund of the Third Institute of Oceanography, SOA (2015035) and the Ocean Welfare Scientific Research Project of China (201105012).
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Fukami H, Chen CA, Chiou CY, Knowlton N. 2007. Novel group I introns encoding a putative homing endonuclease in the mitochondrial cox1 gene of Scleractinian corals. J Mol Evol. 64:591–600.
- Laslett D, Canback B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24;172–175.
- Medina M, Collins AG, Takaoka TL, Kuehl JV, Boore JL. 2006. Naked corals: skeleton loss in Scleractinia. Proc Natl Acad Sci USA. 103:9096–9100.
- Shen KN, Yen TC, Chen CH, Li HY, Chen PL, Hsiao CD. 2014. Next generation sequencing yields the complete mitochondrial genome of the flathead mullet, Mugil cephalus cryptic species NWP2 (Teleostei: Mugilidae). Mitochondrial DNA. 1–2.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Veron JEN, Stafford-Smith M. 2000. Corals of the world. Australian Institute of Marine Science.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20;3252–3255.
