Abstract
The total length of mitogenome of Copsychus saularis is 16 827 bp and contains 13 protein-coding genes, 22 tRNA genes, two rRNA genes and one D-loop. The phylogenetic tree of C. saularis and 13 other species belonging to Passeriformes was built.
Keywords:
The genus Copsychus, containing 12 species, had no mitochondrial genome of them to be determined. The oriental magpie-robin is a common bird of urban gardens and forests that resides in southern Asia (Ali Citation2003; Siddique Citation2008). In this article, we described the mitogenome of C. saularis in order to obtain basic genetic information about this species.
The specimen of C. saularis (Collection number HS15011) was collected from Xiuning County, Anhui Province, China (29.45°N, 118.15°E). The specimen was deposited in the Museum of Huangshan University (Voucher numbers HUM20150020).
The mitogenome of C. saularis (Genbank accession number KU058637) was sequenced to be 16 827 bp which consisted of 13 typical vertebrate protein-coding genes, 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes and one D-loop, which is similar to the typical mtDNA of birds and other vertebrates (Boore Citation1999; Sorenson et al. Citation1999). The overall base composition of the entire genome was as follows: A (29.4%), T (23.3%), C (32.5%) and G (14.8%). All the 13 protein-coding genes initiate with ATG as start codon, except for COI, which begin with GTG. Six of 13 protein-coding genes use the TAA as stop codon, three genes (ND1, COI and ND5) terminate with AGR (AGA and AGG) and the ND6 ends with TAG. The other protein-coding genes (ND2, COIII and ND4) end with the TA- or T- as the incomplete stop codons. Among the mitochondrial protein-coding genes, the ATP8 was the shortest, while the ND5 was the longest. The 22 tRNA genes range in size from 67 to 75 bp. The lengths of 12 s and 16 s rRNA are 982 and 1598 bp. The D-loop in size is 1149 bp.
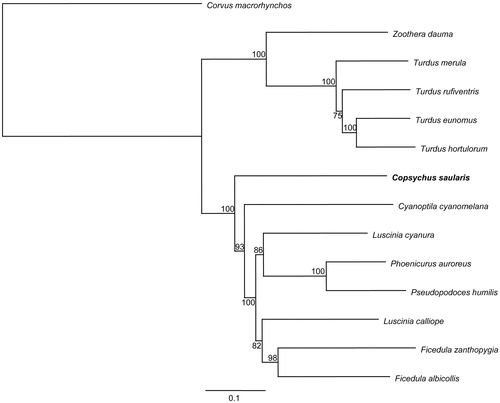
We selected the 12 protein-coding genes of C. saularis and together with other 13 related species to perform phylogenetic analysis. A maximum likelihood tree was constructed by online tool RAxML (Stamatakis et al. Citation2008) (). The phylogenetic analysis result was consistent with the previous research (Sangster et al. Citation2010; Voelker et al. Citation2014).
Figure 1. A maximum likelihood (ML) tree of the 14 species from Passeriformes was constructed based on the dataset of 12 concatenated mitochondrial protein-coding genes by online tool CIPRES. The numbers above the branch meant bootstrap value. Bold black branches highlighted the study species and corresponding phylogenetic classification. The analyzed species and corresponding NCBI accession numbers are as follows: Corvus macrorhynchos (KR072661), Zoothera dauma (KT340629), Turdus merula (KT601060), Turdus rufiventris (KT346357), Turdus eunomus (KM015261), Turdus hortulorum (KF926987), Copsychus saularis (KU058637), Cyanoptila cyanomelana (HQ896033), Luscinia cyanura (KF997864), Phoenicurus auroreus (KF997863), Pseudopodoces humilis (HM535648), Luscinia calliope (HQ690246), Ficedula zanthopygia (JN018411), and Ficedula albicollis (KF293721).
Acknowledgements
We thank Dr Song Huang (Huangshan University) for valuable comments on this manuscript.
Declaration of interest
This project was funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions. The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
References
- Ali S. 2003. The book of Indian birds. Oxford (UK): Oxford University Press. p. 466.
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–80.
- Sangster G, Alström P, Porsmark E, Olsson U. 2010. Multi-locus phylogenetic analysis of Old World chats and flycatchers reveals extensive paraphyly at family, subfamily and genus level (Aves: Muscicapidae). Mol Phylogenet Evol. 57:380–392.
- Siddique YS. 2008. Breeding behavior of Copsychus saularis in Indian-sub-continent: a personal experience. Int J Zool Res. 4:135–137.
- Sorenson MD, Ast JC, Dimcheff DE, Yuri T, Mindell DP. 1999. Primers for a PCR-based approach to mitochondrial genome sequencing in birds and other vertebrates. Mol Phylogenet Evol. 12:105–114.
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web-servers. Syst Biol. 75:758–771.
- Voelker G, Peñalba JV, Huntley JW, Bowie RCK. 2014. Diversification in an Afro-Asian songbird clade reveals founder-event speciation via trans-oceanic dispersals and a southern to northern colonization pattern in Africa. Mol Phylogenet Evol. 73:97–105.
