Abstract
The complete mitochondrial genome of Chionodraco hamatus was obtained, which was 17 457 bp in length. This genome consists of 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes and a putative control region. Of the 37 genes, 28 were encoded by heavy strand, while 9 were encoded by light strand. Overall base composition of mitogenome is estimated to be 26.38% for A, 17.44% for G, 26.00% for T, 30.18% for C, respectively, with a slight A + T bias (52.38%). The phylogenetic analysis based on 13 concatenated protein-coding genes suggested that C. hamatus as a sister species to Chionodraco myersi was clustered in family Chionodraco. The complete mitochondrial genome sequence of C. hamatus could provide a basic data for the studies on evolution for low temperature adaptability, population structure, molecular systematic, stock evaluation and conservation genetics.
Chionodraco hamatus, an important Antarctic fish species of family Channichthyidae, is widely distributed in the High Antarctic zone all around the continent (Kock Citation2005). These fish families have developed unique physiological features that allowed them to survive in the Antarctic seawater that reaches temperatures near to –2 °C. They produce anti-freeze glycoproteins that prevent tissues freezing (Bargelloni et al. Citation1994). Moreover, as an extreme adaptation, the Channichthyidae showed the complete absence of haemoglobin and of functional blood erythrocytes (Ruud Citation1954). Within this family, 11 genera and 15 species are currently recognized, with the large majority of these genera being monospecific. One of the few exceptions is the genus Chionodraco for which three species have been described, namely C. hamatus, Chionodraco myersi and Chionodraco rastrospinosus (Eastman & Eakin Citation2000). But most researchers based their identification primarily on the occurrence and distribution of adults fish (Moteki & Ishimaru Citation2008). In addition, the origin of this group is ambiguous due to the lack of a fossil record although they are thought to have originated from an ancestor of the suborder Percoidei (Buonocore et al. Citation2006). The complete mitochondrial genome sequence of C. hamatus could provide a basic data for the studies on above. So far, no complete mitogenome sequence information of C. hamatus is available.
Adult fish of C. hamatus was collected near Zhongshan Station (68tio 708tio after freezing at −80 °C it was transported to East China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences for storage and DNA extraction. In this study, we obtained the complete mitochondrial genome sequence of C. hamatus, and submitted it into the GenBank database with an accession number KT921282. This mitochondrial genome is 17 457 bp in length and contained 13 protein-coding genes, 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes and a putative control region. Among the 37 genes, 28 were encoded by heavy strand, while 9 were encoded by light strand just as in other teleosts. What differentiated was that the gene arrangement and genome structure were diverse from most teleosts, even from the sister species C. myersi. The NADH dehydrogenase subunit 6 (ND6) gene and the adjacent tRNAGlu were not lost but had been translocated to between tRNAThr and tRNAPro from their canonical location between ND5 and cytochrome b genes. The overall base composition of this genome is 26.38% for A, 17.44% for G, 26.00% for T, 30.18% for C, respectively, with a slight higher A + T content (52.38%).
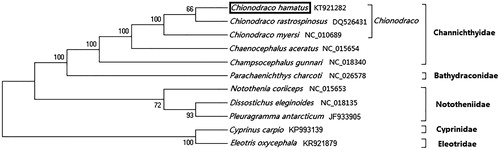
Two kinds of start codons (ATG and GTG) were identified in 13 protein-coding genes; 5 genes ended with TAA, whereas 8 genes had incomplete stop codons TA or T. The length of the total protein-coding genes was 11 444 bp, and the base composition was 23.68% for A, 30.63% for C, 17.63% for G and 28.06% for T. The length of the two rRNA genes was 2639 bp, with the overall base composition of 31.68% for A, 25.54% for C, 21.52% for G and 21.26% for T. The length of 22 tRNA genes was 1593 bp, with the overall base composition of 29.69% for A, 24.67% for C, 21.09% for G, 24.54% for T. The non-coding region was 1810 bp in length, its overall base composition is 28.34% for A, 32.43% for C, 13.48% for G, 25.75% for T. The putative control region was 1349 bp in length, longer than non-Antarctic fishes. Furthermore, only two typical domains including termination-associated sequences (TAS and ETAS) and central conserved sequence block (CSB-E) were detected in the putative control region by comparing with the mitochondrial genomes of other species. The phylogenetic position of C. hamatus within suborder Notothenioidei was reconstructed based on 13 concatenated protein-coding genes using the neighbour-joining method (some mitochondrial genomes without ND6). The phylogenetic tree () shows that C. hamatus first clustered as a monophyletic group with C. rastrospinosus, then together with other three fishes in family Channichthyidae, forming a big branch. Besides, another three fishes in family Nototheniidae formed a big sister branch as well.
Figure 1. Phylogenetic position of Chionodraco hamatus within suborder Notothenioidei based on 13 protein-coding genes using neighbour-joining method. C. hamatus is highlighted with a box. The ND6 was not included in the protein-coding genes of Chionodraco rastrospinosus and Chionodraco myersi analysed here.
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper. This work was supported by the Chinese National Antarctic Research Expedition (CHINARE2014-01-06).
References
- Bargelloni L, Ritchie P, Patarnello T, Battaglia B, Lambert DM, Meyer A. 1994. Molecular evolution at subzero temperatures: mitochondrial and nuclear phylogenies of fishes from Antarctica (suborder Notothenioidei), and the evolution of antifreeze glycopeptides. Mol Biol Evol. 11:854–863.
- Buonocore F, Randelli E, Paderi F, Bird S, Secombes CJ, Mazzini M, Scapigliati G. 2006. The cytokine IL-1 beta from the crocodile icefish Chionodraco hamatus (Perciformes: Channichthyidae). Polar Biol. 29:1018–1027.
- Eastman J, Eakin R. 2000. An updated species list for notothenioid fish (Perciformes; Notothenioidei), with comments on Antarctic species. Arc Fish Mar Res. 48:11–20.
- Kock K-H. 2005. Antarctic icefishes (Channichthyidae): a unique family of fishes. A review, Part I. Polar Biol. 28:862–895.
- Moteki M, Ishimaru T. 2008. Development of feeding and swimming functions in larvae of Chionodraco rastrospinosus (Channichthyidae). Cybium 32:247–251.
- Ruud JT. 1954. Vertebrates without erythrocytes and blood pigment. Nature 173:848–850.
