Abstract
In this study, the complete mitogenome sequence of sea slug, Phyllidia ocellata (Mollusca: Phyllidiidae), has been decoded for the first time by low coverage genome sequencing method. The overall base composition of P. ocellata mitogenome is 31.5% for A, 14.0% for C, 16.4% for G and 38.0% for T, and have low GC content of 30.5%. The assembled mitogenome, consisting of 14 598 bp, has unique 13 protein-coding genes, 22 transfer RNAs and two ribosomal RNAs genes. The P. ocellata mitogenome has the common mitogenome gene organization and feature of Nudipleura (a clade of sea slugs and sea snails). The complete mitogenome of P. ocellata provides essential and important DNA molecular data for further phylogenetic and evolutionary analysis for sea slugs and sea snails.
Sea slugs are marine gastropod mollusks distributed throughout the global oceans. The feeding relationship between the sea slug and the seaweeds, sponges, corals and its special chemical defence system, are the main content and hot point of marine chemical ecology. Phyllidia ocellata is a species of shell-less sea slug, a dorid nudibranch, in the family Phyllidiidae (Bouchet & Rocroi Citation2005). This species is widely distributed throughout the coral reef area of the Indo-West Pacific Ocean, the Red Sea and Australia (Brunckhorst Citation1993; Gosliner et al. Citation2008). This animal can measure up to 60 mm in length, and commonly has a yellow body with a dorsum covered in yellow or white tubercules. There is no gills on the back, is the distinguishing feature of this species. Its mantle contains special glands around the margin which store, as a defensive mechanism, toxic antifeedant chemicals obtained from the sponge prey, so as to establish its chemical defence system, to protect themselves from predators (Jiang et al. Citation2010). Discovery of a large number of bioactive compounds, to suggest that, sea slug is a kind of excellent biomedicine source with high research value.
Samples (voucher no. 394) of P. ocellata were collected from the local aquarium in Taiwan. The methods for genomic DNA extraction, library construction and next-generation sequencing were followed by our previous publication (Shen et al. Citation2014). Initially, the raw next-generation sequencing reads generated from MiSeq (Illumina, San Diego, CA). About 0.02% raw reads (1967 out of 9 288 624) were de novo assembly by using commercial software (Geneious V9, Auckland, New Zealand) to produce a single, circular form of complete mitogenome with about an average 46 X coverage.
The complete mitochondrial genome of P. ocellata was 14 598 bp in size (GenBank KU351090) and its overall base composition is 31.5% for A, 14.0% for C, 16.4% for G and 38.2% for T, and have low GC content of 30.5%, showing 76% identities to the complete mitogenome of Roboastra europaea (GenBank: AY083457) (Grande et al. Citation2002). The protein coding, rRNA and tRNA genes of P. ocellata mitogenome were predicted by using DOGMA (Wyman et al. Citation2004), ARWEN (Laslett & Canbäck Citation2008), MITOS (Bernt et al. Citation2013) tools and manually inspected. The complete mitogenome of P. ocellata includes unique 13 protein-coding genes (PCGs), 22 transfer RNA genes and two ribosomal RNA genes. In the complete mitogenome, four PCGs (ATP8, ATP6, ND3 and COX3), one rRNA (12S rRNA) and eight tRNA (tRNA-Gln, tRNA-Leu 2, tRNA-Asn, tRNA-Arg, tRNA-Glu, tRNA-Met, tRNA-Ser 2 and tRNA-Thr) genes are encoded on L-strand, while other genes are encoded in the H-strand.
The P. ocellata mitogenome has the common gene organization and feature of Nudipleura (a clade of sea slugs and sea snails). It is important to note that six PCGs started with ATG codon (ATP8, COX1, COX2, ND1, ND2 and ND6), three with ATA codon (ATP6, CYTB and ND4), one with ATC codon (ND4L), one with ATT codon (ND3), one with TTG codon (ND5). Seven of 10 PCGs are inferred to terminate with TAA (ATP6, ATP8, COX1, COX2, ND1, ND2, ND3, ND4, ND4L and ND6), other three PCGs with TAG (CYTB, COX3 and ND5) stop codon. The longest one is ND5 gene (1629 bp) in all PCGs, whereas the shortest is ATP8 gene (162 bp). The size of small ribosomal RNA (12S rRNA) and large ribosomal RNA (16S rRNA) genes are 760 and 1127 bp, respectively.
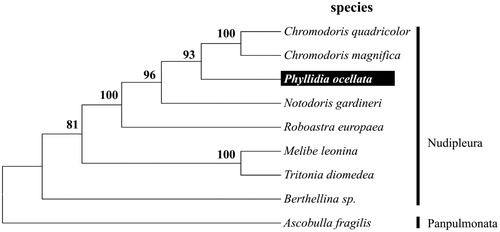
To validate the phylogenetic position of P. ocellata, we used MEGA6 software (Tamura et al. Citation2013) to construct a maximum likelihood tree (with 500 bootstrap replicates) containing complete mitogenomes of eight species derived from Nudibranchia. Ascobulla fragilis derived from Panpulmonata was used as an outgroup for tree rooting. The result shows P. ocellata can be unambiguously grouped in Nudipleura with high bootstrap value supported (). In conclusion, the complete mitogenome of the P. ocellata deduced in this study provides essential and important DNA molecular data for further phylogenetic and evolutionary analysis for sea slugs and sea snails phylogeny.
Figure 1. Molecular phylogeny of Phyllidia ocellata and other related species in Nudipleura based on complete mitogenome. The complete mitogenomes is downloaded from GenBank and the phylogenic tree is constructed by a maximum-likelihood method with 500 bootstrap replicates. The gene’s accession number for tree construction is listed as follows: Ascobulla fragilis (NC_012428), Berthellina sp. (NC_015091), Tritonia diomedea (NC_026988), Melibe leonina (NC_026987), Roboastra europaea (NC_004321), Notodoris gardineri (NC_015111), Phyllidia ocellata (KU351090), Chromodoris magnifica (NC_015096) and Chromodoris quadricolor (KU317089).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Funding information
This research was supported by the Global Climate Change and Ocean Atmosphere Interaction Research, the Marine Biological Sample Museum of the Chinese Offshore Investigation and Assessment, Public Science and Technology Research Funds Projects of Ocean (No. GASI-01-01-04).
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Bouchet P, Rocroi JP. 2005. Classification and nomenclator of gastropod families, Vol 47 (Institute of Malacology). 47:85–397.
- Brunckhorst DJ. 1993. The systematics and phylogeny of Phyllidiid nudibranchs (Doridoidea) (Sydney). 16:1–107.
- Gosliner TM, Behrens DW, Valdés Á. 2008. Indo-Pacific nudibranchs and sea slugs: a field guide to the world’s most diverse fauna. Sea Challengers Natural History Books; California Academy of Sciences. 426 pp.
- Grande C, Templado J, Cervera JL, Zardoya R. 2002. The complete mitochondrial genome of the nudibranch Roboastra europaea (Mollusca: Gastropoda) supports the monophyly of opisthobranchs. Mol Biol Evol. 19:1672–1685.
- Jiang CS, Huang CG, Feng B, Li J, Gong JX, Kurtán T, Guo YW. 2010. Synthesis and antitumor evaluation of methyl spongoate analogs. Steroids 75:1153–1163.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 24:172–175.
- Shen KN, Yen TC, Chen CH, Li HY, Chen PL, Hsiao CD. 2014. Next generation sequencing yields the complete mitochondrial genome of the flathead mullet, Mugil cephalus cryptic species NWP2 (Teleostei: Mugilidae). Mitochondrial DNA. [Epub ahead of print].
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20:3252–3255.
