Abstract
The complete mitochondrial genome of the Oreolalax lichuanensis was determined. It is a circular molecule of 17 702 bp in size and consists of 13 protein-coding genes, 23 tRNA genes, two rRNA genes and a control region (D-loop). The base composition on light strand was 28.0% A, 32.2% T, 24.9% C and 14.9% G. Compared with most other vertebrates, this mitogenome appear a tandem duplication of tRNA-Met gene. This study will facilitate the further research of the population genetics of this species and systematic study of the genus Oreolalax.
Oreolalax lichuanensis (Amphibia, Anura, Megophryidae) is known from southwestern China and it generally occurs in the forest land and at mid-altitudes about 1800 m (Fei et al. Citation2010). We determined the complete mitogenome of this species. The sample of O. lichuanensis was obtained from Hubei Province of China. The GenBank accession number of this mitogenome is KU096847.
The complete mitogenome of O. lichuanensis is 17 Oreolalax lichuanensis 702 bp in length and contains 13 protein-coding genes (PCGs), two ribosomal RNA (rRNA) genes, 23 transfer RNA (tRNA) genes and a control region (D-loop). The base composition of the complete mitochondrial genome was 28.0% A, 32.2% T, 24.9% C and 14.9% G. The A + T base composition (60.2%) was higher than G + C (39.8%), similar to other anurans (Irisarri et al. Citation2010; Chen et al. Citation2011; Shi et al. Citation2012). Except for ND6 gene and eight tRNA genes (tRNA-Gln, Ala, Asn, Cys, Tyr, Ser (UCN), Glu and Pro) encoded on the L-strand, all other genes were encoded on the H-strand. The 12S rRNA (933 bp) and 16S rRNA (1601 bp) genes, locating between tRNA-Phe and tRNA-Leu genes, were separated by tRNA-Val gene. Three types of initiation codons were used for the 13 PCGs, three genes (COI, ND3 and ND5) starting with GTG, one starting with CCT (ND6), and the rest starting with ATG. Four PCGs (ATP6, COIII, ND4 and Cytb) ended with an incomplete stop codon T which may be completed by post-transcriptional polyadenylation with poly A tail. ND5 was the longest gene (1818 bp) and ATP8 gene (168 bp) was the shortest in 13 PCGs. The 23 tRNA genes with the size ranging from 63 bp to 75 bp were interspersed along the whole genome, and most of the tRNAs formed a colver-leaf structure except the second tRNA-Ser, which lost the T ψ C arm. The putative origin of L-strand replication (OL), with a length of 29 bp between the tRNA-Asn and tRNA-Cys genes, can fold into a stem loop of secondary structure, similar to other vertebrates (Chen et al. Citation2011). The D-loop region, which was thought to include the signals for the regulation of mtDNA replication and transcription (Wolstenholme Citation1992), located between tRNA-Trp and tRNA-Phe, was 1701 bp in length. This region is heavily biased to A + T nucleotides (63.5%) and we found some repeated sequences in this region. For the whole mitogenome, there were eight regions of gene overlap (ranging from 1 to 10 bp), and eleven intergenic spacer regions (ranging from 1 to 290 bp).
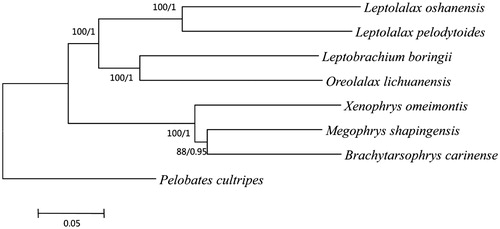
Based on the reported mitogenomes of seven species of Mesobatrachia, four complete mitogenome and three partial mitogenome were used to construct phylogenetic trees. Pelobates cultripes was selected as the outgroup. Maximum-likelihood and Bayesian inference methods yielded the same tree topologies. The phylogram acquired from ML methods are shown in . The article will provide fundamental data for further investigating the phylogenetic study of the species.
Figure 1. The phylogenetic tree inferred by Phyml 3.0 (Guindon et al. Citation2010) and MrBayes v3.2.1 (Ronquist et al. Citation2012). Leptolalax oshanensis (KC460337), Leptolalax pelodytoides (JX564874), Leptobrachium boringii (KJ630505), Xenophrys omeimontis (KP728257), Megophrys shapingensis (JX458090), Brachytarsophrys carinense (JX564854), Pelobates cultripes (NC_008144) and Oreolalax lichuanensis (KU096847).
Funding information
This study was supported by the National Natural Sciences Foundation of China (NSFC–31201702 granted to Bin Wang), the Key Laboratory of Mountain Ecological Restoration and Bioresource Utilization of CIB, CAS, the Ecological Restoration and Biodiversity Conservation Key Laboratory of Sichuan Province.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Chen GY, Wang B, Liu JY, Xie F, Jiang JP. 2011. Complete mitochondrial genome of Nanorana pleskei (Amphibia: Anura: Dicroglossidae) and evolutionary characteristics of the amphibian mitochondrial genomes. Curr Zool. 57:785–805.
- Fei L, Ye CY, Jiang JP. 2010. Colored atlas of Chinese amphibians. Chengdu: Sichuan Publishing House of Science and Technology.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59:307–321.
- Irisarri I, San Mauro D, Green DM, Zardoya R. 2010. The complete mitochondrial genome of the relict frog Leiopelma archeyi: insights into the root of the frog Tree of Life. Mitochondr DNA. 21:173–182.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Shi G, Jin X, Zhao S, Xu T, Wang R. 2012. Complete mitochondrial genome of Trypauchen vagina (Perciformes, Gobioidei). Mitochondrial DNA. 23:151–153.
- Wolstenholme DR. 1992. Animal mitochondrial DNA: Structure and evolution. International Review of Cytology. 141:173–216.
