Abstract
In our study, we first report complete mitogenome for P. ruficollis and obtain basic genetic information. The genome of P. ruficollis is 17 009 bp which contained 13 protein-coding genes, 22 transfer RNA genes, two ribosomal RNA genes and two control regions. Overall bases composition of the complete mitochondrial genome is 29.70%A, 14.47%G, 23.31% T, 32.52%C. Twelve PCGs and 14 tRNA genes are distributed on the H-strand, ND6 and eight tRNA genes are encoded on the L-strand.
Pomatorhinus ruficollis (Rufous-necked scimitar babbler) is one of the members in Pomatorhinus genus, they distributed in northern Indochina, northern and western Burma, central to southern China and sino-Himalayan. Pomatorhinus rufillos are mainly live in lowlands, foothills and montane forests (Nyari & Reddy Citation2013) range altitude from 200 m to 3000 m (Song et al. Citation2011). The population of P. ruficollis sudden drop due to e-waste polluted (Zhang et al. Citation2015). Because P. ruficollis chest differ in the colours spots, vertical lines and it can be slipted into at least three groups (Cheng Citation1964; Collar Citation2006; Collar & Robson Citation2007). Based on the morphological characteristics of P. rufillos includes 14 subspecies (Cheng Citation1962). P.r. musicus is promoted to be independent specie with morphometric differences (Collar Citation2004, Citation2006). Therefore, P. ruficollis contains 13 subspecies (Song et al. Citation2011). The complete mitochondrial genome of P.ruficollis will be helpful for the deeply study on the genetic phylogeny and can provided the method for species conservation.
In our experiment, the muscle sample of P. ruficollis was collected from Ya'an (N30°01', E103°02') and stored at the Wildlife Conservation Laboratory, Sichuan Agricultural University, Sichuan province, China. The mitochondrial genome extracted from the muscle tissue by phenol-chloroform method (Sambrook & Russell Citation2001). We designed 21 pairs of normal PCR primers and four pairs LA-PCR primers by primer 5.0 according to complete mitochondrial genome of Garrulax cineraceus (GeneBank accession no. NC024553). MAGE6.0 (Tamura et al., 2013) is used to construct neighbour-joining (NJ) tree.
The complete mitogenome of P. ruficollis is 17 009 bp submitted to GenBank (KT970675). The mitogenome have 13 protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes and two control regions (D-loop). Twelve PCGs and 14 tRNA are distributed on the H-strand, ND6 and eight tRNA genes are encoded on the L-strand. The base composition of mitogenome is 29.70%A, 14.47%G, 23.31% T and 32.52%C, so the percentage of A + T (53.01%) is slightly higher than G + C (46.99%). Twelve of the 13 protein-coding genes regard ATG as start codon, except GTG for ND1 gene. Four PCGs (ND2, COX3, ND3 and ND4) showed incomplete stop codons (T or TA), the five genes (COX2, ATP8, ATP6, ND4 and Cytb) end with TAA, ND1 and ND5 stops with AGA, COX1 use AGG, and ND6 terminated with TGA. Twelve SrRNA and 16 SrRNA are located between tRNAPhe and tRNALeu(UUR) that is separated by tRNAVal, with the size of 982 and 1601 bp, respectively. The two control regions (CR) of the P. ruficollis are 1058 bp and 385 bp, CR1 is located between the tRNAThr and tRNAPro and CR2 between tRNAGlu and tRNAPhe. Pomatorhinus ruficollis complete mitochondrial genome structure is similar to the typical mitochondrial genomse of vertebrates (Qian et al. Citation2012; Zhang et al. Citation2013; Duan et al. Citation2014).
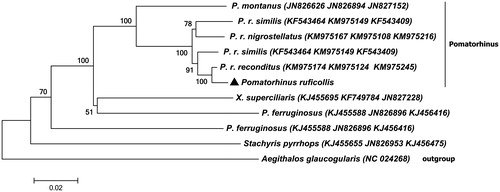
shows that Pomatorhinus genus gathered to one clade, Xiphirhynchus superciliaris and Pomatorhinus ferruginosus are clustered as a sister lineage. Pomatorhinus is not monophyly, which is consistent with other studies (Cibois Citation2003; Collar Citation2004; Reddy & Moyle Citation2010; Moyle et al. Citation2012).
Figure 1. The phylogenetic tree used the method of neighbour-joining (NJ) tree based on ND2, ND3, Cytb protein-coding genes with Kimura 2-parameter model by using MEGA 6.0 program (MEGA Inc., Englewood, NJ). The number on each branch indicated the local bootstrap value. The ND2, ND3 and Cytb Genebank accession number are arranged in brackets after the name of the species.
Disclosure statement
This work was supported by the National Natural Science Foundation of China (31370407). The authors report no conflicts of interest, and are alone responsible for the content and writing of the paper.
References
- Collar NJ. 2004. Species limits in some Indonesian thrushes. Forktail. 20:71–87.
- Collar NJ. 2006. A partial revision of the Asian babblers (Timaliidae). Forktail. 22:85–112.
- Collar NJ, Robson C. 2007. Family Timaliidae (Babblers). In: del Hoyo J, Elliott A, Christie DA, editors. Handbook of the birds of the world, vol. 12. Barcelona: Lynx Edicions. p. 70–291.
- Cibois A. 2003. Mitochondrial DNA phylogeny of babblers(Timaliidae). Auk. 120:35–54.
- Cheng TH. 1962. A systematic review of Pomatorhinus heretofore recorded from China. Acta Zool Sin. 14:197–218.
- Cheng TH. 1964. The Chinese birds retrieval system. Beijing: Science Press.
- Duan X, Zhang H, Li H, Niu L, Wang L, Li L, Zhong T. 2016. Genetic characterization of Meigu goat (Capra hircus) based on the mitochondrial DNA. Mitochondrial DNA. 27:1533–1534.
- Moyle RJ, Andersen MJ, Olivero CJ. 2012. Phylogeny and biogeography of the Core Babblers (Aves: Timaliidae). Syst Biol. 61:631–651.
- Nyari A, Reddy S. 2013. Comparative phyloclimatic analysis and evolution of ecological niches in the Scimitar Babblers (Aves:Timaliidae:Pomatorhinus). PLoS One. 8:e55629.
- Qian C, Wang Y, Guo Z, Yang J, Kan X. 2013. Complete mitochondrial genome of Skylark, Alauda arvensis (Aves: Passeriformes): The first representative of the family Alaudidae with two extensive heteroplasmic control regions. Mitochondrial DNA. 24:246–248.
- Reddy S, Moyle RG, 2010. Systematics of the scimitar babblers (Pomatorhinus:Timaliidae): phylogeny, biogeography, and species-limits of four species complexes. Biol J Linnean Soc 102:846–869.
- Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, Vol. 6, 3rd ed. New York: Cold Spring Harbor Laboratory Press. p. 4–6.
- Song R, Dong F, Liu LM, Wu F, Wang K, Zou FS, Yang XJ. 2011. Preliminary discussion on the phylogenetic and taxonomic relationship of Pomatorhinus ruficollis. Zool Res. 32:241–247.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol, mst197.
- Zhang H, Li Y, Wu X, Xue H, Yan P, Wu XB. 2015. The complete mitochondrial genome of Paradoxornis webbianus (Passeriformes, Muscicapidae). Mitochondrial DNA. 26:879–880.
- Zhang Q, Wu J, Sun Y, Zhang M, Mai B, Mo L, et al. 2015. Do bird assemblages predict susceptibility by e-waste pollution? A comparative study based on species and guild-dependent responses in China Agroecosystems. PLoS One. 10:e0122264.
