Abstract
The complete mitochondrial genome of Papilio xuthus (Lepidoptera: Papilionidae) was determined in this study. The mitochondrial genome is a circular molecule of 15 359 and contains 37 genes including 13 protein-coding genes (PCGs), 22 transfer RNA genes, two ribosomal RNA genes and one control region. The nucleotide composition of the A. chinensis mitogenome is strongly biased toward A + T nucleotides (80.45%). Nine protein-coding genes and 14 tRNA genes are encoded on the H strand, and the other four protein-coding genes and eight tRNA genes are encoded on the L strand. The gene order and the orientation of their mitogenomes were similar to all know Papilionidae species. Finally, the phylogenetic relationships of 11 Papilionidae species were reconstructed based on complete mitochondrial genome using the Bayesian inference (BI) and the maximum-likelihood (ML) method. These molecular-based phylogenies support the traditional morphologically based view of relationships within the Papilionidae.
The papilionid butterflies, including about 530 species, are distributed worldwide (Yagi et al. Citation1999). As many of them have large beautiful wings, have greatly contributed to studies of ecology, behavior and evolution in insects (Scriber Citation1995; Boggs et al. Citation2003). The butterfly Popilio xuthus mainly distributed in East Asia (Li et al. Citation2015) and the larva is a famous pest of agriculture (Kong et al. Citation2006). The specimen was collected in the Shengjin Lake National Nature Reserve, Anhui province, China, in August 2015. Total genomic DNA was extracted from muscle tissue using the standard phenol–chloroform protocol (Sambrook et al. Citation2001). The mitochondrial genome was amplified by polymerase chain reaction (PCR) using five pairs of primers. Except for the A + T-rich region, the PCR fragments were directly sequenced by primer walking. The A + T-rich region of lepidopterans contains several features that obscure proper base calling, so the PCR production was sequenced after cloning.
The mitochondrial genome is a circular molecule of 15 359 bp (accession no. KU356933) and contains 37 genes including 13 protein-coding genes (PCGs), 22 transfer RNA genes, two ribosomal RNA genes and one control region. Among these, 14 genes were encoded on the L-strand, including four PCGs (ND1, ND4, ND4L and ND5), two rRNA genes, eight tRNA genes (tRNAGlu, tRNACys, tRNATyr, tRNAPhe, tRNAHis, tRNAPro, tRNAleu(CUN) and tRNAVal) and A + T-rich region. The remaining 23 genes are encoded on the H strand. The arrangement of genes is similar to all know Papilionidae species.
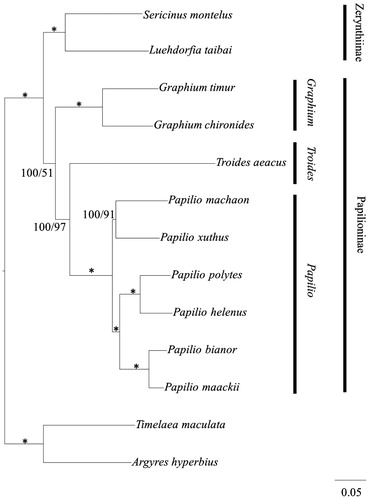
Eleven Papilionidae species were used to reconstruct phylogenetic tree based on the complete mitochondrial genome through the Bayesian inference (BI) and maximum-likelihood (ML) methods, using Timelaea maculate and Argyres hyperbius as the outgroup. MrBayes Version 3.1.2 (Ronquist & Huelsenbeck Citation2003) and a PHYML online web server (Guindon & Gascuel Citation2003; Guindon et al. Citation2005) were employed to reconstruct BI and ML trees, respectively. For BI analysis, the model GTR + I+G was selected via MrModeltest version 2.1 (Nylander et al. Citation2004), the MCMC was run for 1 000 000 generations until the average standard deviation of split frequencies reached a value less than 0.01, with the Bayesian posterior probabilities calculated from the sample points after the MCMC algorithm had started to converge (Zhan & Fu Citation2011). The result of phylogenetic tree among the 11 Papilionidae species is shown in . It is clearly seen that the phylogenetic tree is divided into two major clades. The first lineage, subfamily Papilioninae, includes species of genus Papilio (Papilio bianor, Papilio maackii, Papilio machaon, P. xuthus, Papilio polytes and Papilio helenus), Graphium (Graphium chironides and Graphium timur), Troides (Troides aeacus). The subfamily Zerynthiinae (Sericinus montela and Luehdorfia taibai) forms the second clade and is sister to subfamily Papilioninae. These molecular-based phylogenies support the traditional morphologically based view of relationships within the Papilionidae (Wu et al. Citation2001).
Figure 1. Inferred phylogenetic relationships among Papilionidae based on the complete mitochondrial genome using Bayesian inference (BI) and maximum-likelihood (ML) analysis. Branch lengths and topologies came from the Bayesian analyses. Numbers in each branch indicate Bayesian posterior probabilities (BPP)/maximum-likelihood (ML) bootstrap values. *Indicates posterior probabilities = 100 and ML bootstrap = 100. GenBank accession numbers for the published sequences are NC018040.1 (Papilio bianor), NC021411.1 (Papilio maackii), NC018047.1 (Papilio machaon), KU356933 (Papilio xuthus), NC024742.1 (Papilio polytes), NC026910.1 (Graphium chironides), NC024098.1 (Graphium timur), EU625344.1 (Troides aeacus), HQ259122.1 (Sericinus montela), NC023938.1 (Luehdorfia taibai), NC025757.1 (Papilio helenus), NC021090.1 (Timelaea maculate), NC015988.1 (Argynnis hyperbius).
Our study of P. xuthus can provide a useful database for analyzing the classification and status in Papilionidae. In addition, it is useful to construct molecular identification of this species.
Acknowledgements
The authors thank Shuyan Wang, Na Li, Chunqin Sun and Pengyu Yang for helpful experiment, corrections and comments regarding this manuscript.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Funding information
This project was supported by the Foundation for Projects in the National Science & Technology Pillar Program during the Twelfth Five-year Plan Period (No. 2012BAK11B03) and the Foundation for Graduate Student Academic Innovation Research Project of Anhui University (No. yqh100135).
References
- Boggs CL, Watt WB, Ehrlich PR. 2003. Butterflies: ecology and evolution taking flight, Chicago: University of Chicago Press.
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52:696–704.
- Guindon S, Lethiec F, Duroux P, Gascuel O. 2005. PHYML Online – a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 33:W557–W559.
- Kong HR, Liang XC, Luo YZ. 2006. Anatomy of the reproductive system of Papilio xuthu Linnaeus. J – Yunnan Agric Univ. 21:459–462.
- Li X, Fan D, Zhang W, Liu G, Zhang L, Zhao L, Fang X, Chen L, Dong Y, Chen Y. 2015. Outbred genome sequencing and CRISPR/Cas9 gene editing in butterflies. Nat Commun. 1–10
- Nylander JA, Ronquist F, Huelsenbeck JP, Nieves-Aldrey J. 2004. Bayesian phylogenetic analysis of combined data. Syst Biol. 53:47–67.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Scriber JM. 1995. Overview of swallowtail butterflies: taxonomic and distributional latitude. In: Scriber JM, Tsubaki Y, Lederhouse RC. (Eds.), Swallowtail Butterflies: Their Ecology and Evolutionary Biology. Gainesville, FL: Scientific Publishers. pp. 3–20.
- Sambrook JM, Russell DW, Janssen K, Argentine J. 2001. Molecular cloning: a laboratory manual on the web. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
- Wu C, Liu Q, Lu L, Gong Z, Wang D, Yu X, Li C, Liu J. 2001. Fauna sinica. Insecta. 25:241–245.
- Yagi T, Sasaki G, Takebe H. 1999. Phylogeny of Japanese papilionid butterflies inferred from nucleotide sequences of the mitochondrial ND5 gene. J Mol Evol. 48:42–48.
- Zhan A, Fu J. 2011. Past and present: phylogeography of the Bufo gargarizans species complex inferred from multi-loci allele sequence and frequency data. Mol Phylogenet Evol. 61:136–148.
