Abstract
The flower chafer Osmoderma opicum (Coleoptera: Scarabaeidae) has been listed in Korea as a class II endangered wild species. The 15 341-bp complete mitochondrial genome (mitogenome) of the species consists of a typical set of genes (13 protein-coding genes [PCGs], two rRNA genes and 22 tRNA genes) and the A + T-rich region, with an arrangement typical of insects. The 761-bp long A + T-rich region of O. opicum has a microsatellite-like sequence consisting of (TA)6, along with a poly-T (12 bp) and a poly-A (16 bp) stretch, but does not have a longer repeat sequence often found in other Scarabaeidae. Twelve PCGs started with the typical ATN codon, whereas the COI started with AAC, which has been designated as the start codon for the coleopteran suborder Polyphaga. Phylogenetic analysis using 11 PCGs from Scarabaeidae showed that O. opicum was placed as the sister to the within-subfamilial species Myodermum sp., forming the monophyletic group, the subfamily Cetoniinae, with moderate to high nodal support.
The flower chafer Osmoderma opicum (Coleoptera: Scarabaeidae) is distributed throughout Korea and Japan (Won et al. Citation1998; Bayartogtokh et al. Citation2012). In Korea, it is found yearly in the north-eastern mountainous areas, from July to August, but with rarity, and thus, is listed as a class II endangered wild animal in Korea (Won et al. Citation1998). In this study, we sequenced the complete mitochondrial genome (mitogenome) of O. opicum to accumulate genetic information of the species. After obtaining the necessary permissions, one adult was collected from a mountain located in Yeongju City, Gyeongsangbuk-do Province, Korea (a detailed description of the administrative district including coordinates is omitted). A voucher specimen was deposited in the National Institute of Biological Resources, Incheon, Korea.
By using the total DNA as template, two long fragments (COI–CytB and CytB–COI) were amplified using PCR primers designed in this study (data not shown), and subsequently, 30 short fragments were amplified using the long fragments as templates and the primers designed in this study (data not shown).
The O. opicum mitogenome is 15 341 bp in length and includes sets of genes (two rRNAs, 22 tRNAs and 13 protein-coding genes [PCGs]) and a major non-coding 761 bp A + T-rich region (GenBank accession no. KU500641) with arrangements typically found in majority of insects. Currently, a total of 16 mitogenomes are reported from Scarabaeidae, but only three of them are complete with the sequences of A + T-rich region ranging in sizes from 2590 to 5654 bp. The extraordinarily long A + T-rich regions of these species stem either from the presence of seven 82-bp and twenty-eight 117-bp long tandem repeats separated by non-repeat sequences in Protaetia brevitarsis (Kim et al. Citation2014) and ten 88-bp and two 153-bp long tandem repeats abutting each other in Cheirotonus jansoni (Shao et al. Citation2014) or no major repeat units but the presence of a small microsatellite region consisting of TA in Rhopaea magnicornis (Cameron et al. Citation2009). The A + T-rich region of O. opicum in the present study is similar to that of R. magnicornis in that it only has a microsatellite-like sequence consisting of (TA)6, along with a poly-T (12 bp) and a poly-A (16 bp) stretch. Twelve PCGs started with the typical ATN codon (four with ATG, three with ATT and five with ATA), whereas the COI gene started with AAC, designated as the start codon for the coleopteran suborder Polyphaga (Sheffield et al. Citation2008).
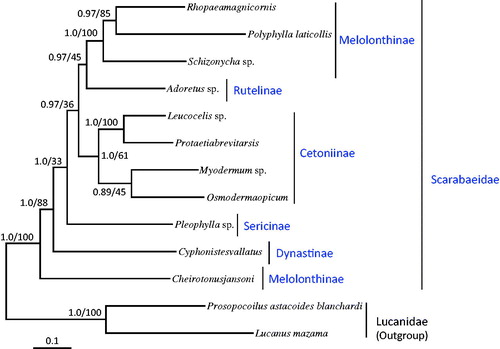
We downloaded the available mitogenome sequences of 10 Scarabaeidae from GenBank along with two species of Lucanidae for use as outgroups and 11 (excluding ND1 and ND2 because of unavailability) among 13 PCGs for phylogenetic analysis. Maximum-likelihood (ML) and Bayesian inference (BI) methods were performed using the GTR + I+G model in CIPRES Portal v. 3.1 (Miller et al. Citation2010). Both the analytical methods presented identical topologies with O. opicum as a sister to the within-subfamilial species Myodermum sp., but the nodal support for this group was moderate to low (ML, 45%; BI, 0.89) (). The subfamily Cetoniinae, in which O. opicum is included, formed a strong monophyletic group in the BI analysis (1.0) but was low in the ML method (61%).
Figure 1. Phylogeny of Scarabaeidae. Bayesian inference (BI) and maximum likelihood (ML) methods produced the same topology based on the concatenated 11 protein-coding genes (PCGs). ND1 and ND2 were excluded in the analysis because of unavailability in several other species of Scarabaeidae. The numbers at each node specify Bayesian posterior probabilities in percent by BI (first value) and bootstrap percentages of 1000 pseudoreplicates by ML (second value). The scale bar indicates the number of substitutions per site. Two species of Lucanidae were utilized as outgroups. GenBank accession numbers are as follows: Rhopaea magnicornis, FJ859903; Polyphylla laticollis, KF544959; Schizonycha sp., JX412739; Adoretus sp., JX412788; Leucocelis sp., JX412740; Protaetia brevitarsis, KJ830749; Myodermum sp., JX412847; Pleophylla sp., JX412736; Cyphonistes vallatus, JX412731; Cheirotonus jansoni, KC428100; Prosopocoilus astacoides blanchardi, KF364622; and Lucanus mazama, NC_013578.
Disclosure statement
This study was supported by a grant from the National Institute of Biological Resources (NIBR), funded by the Ministry of Environment (MOE) of the Republic of Korea (NIBR201503103).
References
- Bayartogtokh B, Kim JI, Bae YJ. 2012. Lamellicorn beetles (Coleoptera: Scarabaeoidea) in Korea and Mongolia. Entomol Res. 42:211–218.
- Cameron SL, Sullivan J, Song H, Miller K.B, Whiting MF. 2009. A mitochondrial genome phylogeny of the Neuropterida (lace-wings, alderflies and snakeflies) and their relationship to the other holometabolous insect orders. Zool Scr. 38:575–590.
- Kim MJ, Im HH, Lee KY, Han YS, Kim I. 2014. Complete mitochondrial genome of the whiter-spotted flower chafer, Protaetia brevitarsis (Coleoptera: Scarabaeidae), Mitochondrial DNA. 25:177–178
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans (LO). p. 1–8.
- Shao LL, Huang DY, Sun XY, Hao JS, Cheng CH, Zhang W, Yang Q. 2014. Complete mitochondrial genome sequence of Cheirotonus jansoni (Coleoptera: Scarabaeidae). Genet Mol Res. 13:1047–1058.
- Sheffield NC, Song H, Cameron SL, Whiting MF. 2008. A comparative analysis of mitochondrial genomes in Coleoptera (Arthropoda: Insecta) and genome descriptions of six new beetles. Mol Biol Evol. 25:2499–2509.
- Won BH, Kwon YJ, Kim SS, Kim W, Kim IS, Kim JH, Kim JI, Nam SH, Rho BJ, Moon TY, et al. 1998. Endangered wild species in Korea. Seoul, Korea: Kyo-Hak Publishing Co.
