Abstract
The complete mitochondrial genome was determined from a Microhyla butleri, Microhylidae, Microhyla, which was collected from Shenzhen, China. The mitogenome was 16 714 bp in length, containing 13 protein-coding genes, two rRNA genes, 22 tRNA genes and a control region (D-loop). The base composition was 28.7% A, 29.5% T, 27.2% C and 14.6% G. The gene order and contents were identical to most amphibian mitogenome. Except ND1 gene beginning with GTG and COI gene beginning with ATA, all other protein-coding genes began with ATG as start codon. Six protein-coding genes (ND1, COII, ATP6, COIII, ND3 and ND4) ended with incomplete stop codon T. The 22 tRNA genes with the size ranging from 65 bp to 74 bp were interspersed along the whole genome. The D-loop region containing tandem repetition was 1334 bp in length and heavily biased to A + T nucleotides.
Microhyla butleri is known from central, southern and south-western China and it generally occurs in lowlands and at mid-altitudes up to about 220–1500 m (Frost Citation2015). We determined the complete mitogenome of this species. The sample of M. butleri was obtained from Shenzhen, Guangdong Province of China, with voucher number CIB20150408. Its muscle tissue sample was fixed with 95% ethanol and stored at −20 °C in the herpetological collection at Chengdu Institute of Biology, Chinese Academy of Sciences. The GenBank accession number is KT972718.
At present, four mitochondrial genome sequences of Microhyla species have been reported, including M. pulchra, M. okinavensis, M. heymonsi and M. ornata (GenBank accession numbers:NC_024547, AB303950, NC_006406 and NC_009422) (Zhang et al. Citation2005; Igawa et al. Citation2008; Wu et al. Citation2016). It is difficult to distinguish M. ornata, M. heymonsi and M. butleri from the appearance. In our study, we used 10 pairs of specific primers to amplify the mitochondrial complete genome, expecting to provide preliminary molecular data for further research.
The gene order and structure of M. butleri mitogenome is a circular molecule of 16 714 bp in size, containing 13 protein-coding genes, two rRNA genes, 22 tRNA genes and a control region. The base composition of the complete mitochondrial genome was 28.7% A, 29.5% T, 27.2% C and 14.6% G. The A + T base composition (58.2%) was higher than G + C (41.8%), agreeing with most vertebrates. The arrangement of genes in this mitogenome is identical to the general order of amphibian mitogenome (Chen et al. Citation2011). ATP8 gene (165 bp) was the shortest and ND5 was the longest gene (1791 bp) in 13 protein-coding genes. All protein-coding genes use ATG as start codon except ND1 gene beginning with GTG and COI gene beginning with ATA. Except ND6 gene and eight tRNA genes (tRNA-Pro, tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-Ser and tRNA-Glu) encoded on L-strand, remaining genes were coded on the heavy strand (H-strand). Six genes (ND1, COII, ATP6, COIII, ND3 and ND4L) ended with an incomplete stop codon T which may be completed by posttranscriptional polyadenylation with poly A tail. The 12S rRNA (941 bp) and 16S rRNA (1579 bp) genes, located between tRNA-Phe and tRNA-Leu genes, were separated by tRNA-Val gene. The 22 tRNA genes with the size ranging from 65 bp to 74 bp were interspersed along the whole genome, and most of the tRNAs formed a colver-leaf structure except tRNA-Ser, which lost the T ψ C arm. The putative origin of L-strand replication (OL), with a length of 31 bp between the tRNA-Asn and tRNA-Cys genes, can fold into a stem loop of secondary structure, similar to other vertebrates (Boore Citation1999). The D-loop region, located between Cytb and tRNA-Leu, was 1333 bp in length. This region is heavily biased to A + T nucleotides (61.05%) and we found some repeated sequences in this region. For the whole mitogenome, there were 11 regions of gene overlap (ranging from 1 to 10 bp) and 11 intergenic spacer regions (ranging from 1 to 32 bp).
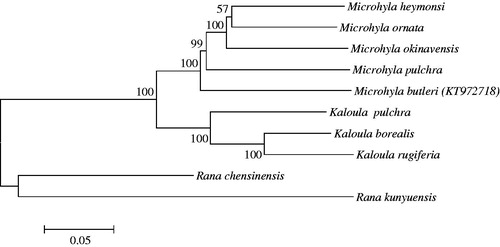
Based on the published mitogenome sequences of Microhylidae species, Rana chensinensis and Rana kunyuensis were selected as outgroup taxa to reconstruct a phylogenetic tree () using MEGA 6.0 (Tamura et al. Citation2013). Bayesian inference (BI) and maximum-likelihood (ML) analyses were used to reconstruct phylogenetic tree, which yielded the same topologies (). The article will provide fundamental data for further investigating the phylogenetic study of the species.
Figure 1. Neighbour-joining tree using MEGA 6.0 based on complete mitochondrial genomes. Microhyla heymonsi (NC_006406), Microhyla ornata (NC_009422), Microhyla okinavensis (AB303950), Microhyla pulchra (NC_024547), Microhylabutleri (KT972718), Kaloula pulchra (AY458595), Kaloula borealis (NC_020044), Kaloula rugiferia (KP682314), Rana chensinensis(NC_023529) and Rana kunyuensis (KF840516).
Funding information
This study was supported by the National Natural Sciences Foundation of China (31471964) and Important Research Project of Chinese Academy of Sciences (KJZG-EW-L13), the Key Laboratory of Mountain Ecological Restoration and Bioresource Utilization of CIB, CAS, the Ecological Restoration and Biodiversity Conservation Key Laboratory of Sichuan Province.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Chen GY, Wang B, Liu JY, Xie F, Jiang JP. 2011. Complete mitochondrial genome of Nanorana pleskei (Amphibia: Anura: Dicroglossidae) and evolutionary characteristics of the amphibian mitochondrial genomes. Curr Zool. 57:785–805.
- Frost DR. 2015. Amphibian species of the world: an online reference. Version 6.0. American New York (NY): Museum of Natural History. Available from: http://research.amnh.org/herpetology/amphibia/index.html (Accessed 16 November 2015).
- Igawa T, Kurabayashi A, Usuki C, Fujii T, Sumida M. 2008. Complete mitochondrial genomes of three neobatrachian anurans: a case study of divergence time estimation using different data and calibration settings. Gene. 407:116–129.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wu XY, Li YM, Zhang HB,Yan L,W XB. 2016. The complete mitochondrial genome of Microhyla pulchra (Amphidia, Anura, Microhylidae). Mitochondrial DNA. 27:40–41.
- Zhang P, Zhou H, Chen YQ, Liu YF, Qu LH. 2005. Mitogenomic perspectives on the origin and phylogeny of living amphibians. Syst Biol. 54:391–400.
