Abstract
The Barbodes hexagonolepis, as an important commercial species, had the limited resources on biology and genetics. We reported the complete mitogenome of this species, and got a whole length of 16 587 bp (containing two rRNAs, 22 tRNAs, 13 mRNAs and one D-loop region). The two rRNAs located between tRNAphe and tRNAleu and were separated by tRNAval. The 22 tRNAs was ranged from 66 bp (tRNAcys) to 76 bp (tRNAleu and tRNAlys) in length. The 13 mRNAs had four overlap regions, three types of start codons and four types of termination codons. The D-loop region was found between tRNAPro and tRNAPhe. To further explore the phylogenetic relationship of the B. hexagonolepis, we constructed the phylogenetic tree and verified that the B. hexagonolepi was a part of the Barbinae and independent from other genus of the Barbinae. This result provided the valuable evidence on phylogenetic relationship of the B. hexagonolepi at the molecular level and essential resource for further research on this species.
The Barbodes hexagonolepis, a species of genus Barbodes in Cyprinidae, located in Brahmaputra and the Irrawaddy basin (Nelson Citation2006). This freshwater fish was distinguished by its mouth (horizontal gape in male and acclivitous in female) and recorded in 1989 (Chen et al. Citation1999). The B. hexagonolepis had a significant dorsal ridge and a deep furcate tail fin (Froese & Pauly Citation2000). Because of the succulent and delicious fish, the B. hexagonolepis was treated as the valuable commercial species and local popular fish. Nevertheless, the G. kempi was less popularity, which owing to the limited resource on distribution, biology and genetics of this species, including mitochondrion.
The fin of the B. hexagonolepis was sampled from lower reach of Brahmaputra (Motuo, Tibet). The genomic DNA was extracted by the DNeasy Tissue Kit (Qiagen, Hilden, Germany) and the complete mitochondrion was amplified by distinct primers and assembled by DNAstar v7.1 (DNASTAR, Inc., Madison, WI), following the standard procedure. The complete mitochondrion of the B. hexagonolepis was 16 587 bp in length (GenBank accession no. KU380329), including two rRNAs (12S and 16S rRNA), 22 tRNAs, 13 mRNAs and one D-loop region.
The 12S rRNA and the 16S rRNA were 956 bp and 1679 bp in length respectively, and the two rRNAs located between tRNAphe and tRNAleu and were separated by tRNAval. The 22 tRNAs was ranged from 66 bp (tRNAcys) to 76 bp (tRNAleu and tRNAlys) in length, and distributed through the whole mitochondrion. Among the 13 mRNAs, the longest was ND5 (1824 bp in length) and the shortest was ATP8 (165 bp in length). There existed four overlap regions among mRNAs (ATP8 and ATP6 with 7 bp, ATP6 and COX3 with 1 bp, ND4L and ND4 with 7 bp, ND5 and ND6 with 4 bp). These 13 mRNAs had three types of start codons and four types of termination codons. The start codons were “ATG” (ND1, ND2, COX2, ATP8, COX3, ND3, ND4L, ND4, ND5 and CYTB), “GTG” (COX1 and ATP6) and “TTA” (ND6). The termination codons were “TAA” (ND1, COX1, ATP6, ND4L and ND5), “TAG” (ND2, ATP8 and ND3), “T–” (COX2, CO3, ND4 and CYTB) and “CAT” (ND6). Furthermore, the D-loop region (921 bp in length) was found between tRNAPro and tRNAPhe.
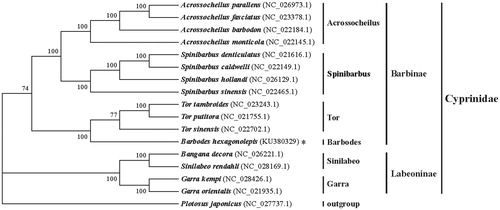
The Barbodes is one genus of subfamily Barbinae, contained in the family Cyprinidae (Chen & Chen Citation2001). To explore the phylogenetic relationship of the B. hexagonolepis among other species of the Barbinae and the Labeoninae (these two subfamilies belong to Cyprinidae), the complete mitochondrion sequences of other 15 species of five genus from the Cyprinidae (mainly from Barbinae and the Labeoninae) and the complete mitochondrion sequence of the B. hexagonolepi in this study were used to construct the phylogenetic tree by MEGA6.06 software (MEGA Inc., Englewood, NJ) with ML analysis, and took the Plotosus japonicas as the outgroup (). The neighbour-joining (NJ) tree (with 1000 bootstrap replicates) and the perfect bootstrap of each cluster verified that the B. hexagonolepi was a part of the Barbinae and independent from other genus of the Barbinae, such as the Acrossocheilus, the Spinibarbus and Tor. This result provided the valuable evidence on phylogenetic relationship of the B. hexagonolepi at the molecular level and essential resource for further research on this species.
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of the article. This study was supported by the National “Twelfth Five-Year” Plan for Science & Technology Support of China (2012BAD25B) and the Fundamental Research Funds for the Central Universities (XDJK2016C047).
References
- Chen X-Y, Yang J-X, Chen Y-R. 1999. A review of the cyprinoid fish genus Barbodes Bleeker, 1859, from Yunnan, China, with descriptions of two new species. Zool Studies – Taipei. 38:82–88.
- Chen Z-M, Chen Y-F. 2001. Phylogeny of the specialized schizothoracine fishes (Teleostei: Cypriniformes: Cyprinidae). Zool Studies – Taipei. 40:147–157.
- Froese R, Pauly D. 2000. FishBase 2000: concepts, design and data sources (WorldFish).
- Nelson JS. 2006. Fishes of the world. New York: John Wiley & Sons.