Abstract
The gray-headed flying squirrel Petaurista elegans caniceps is a cryptic mammal which is not well known. Our study sequenced the complete mitochondrial genome of Petaurista elegans caniceps. The mitochondrial genome is 16 520 bp in size, including 22 tRNA genes, 13 protein-coding genes, 12S rRNA, 16S rRNA and 1 control region. Phylogenetic analysis was performed using whole mitogenome sequences with other 14 closely related taxa to assess their phylogenetic relationship. This mitochondrial genome sequence will provide a better understanding for P. elegans caniceps evolution in the future.
The gray-headed flying squirrel Petaurista elegans caniceps belongs to the subfamily Sciurinae and is distributed mainly in the southwest of China, Nepal and Burma (Smith & Xie Citation2008). P. elegans caniceps is an arboreal and nocturnal mammal (Smith & Xie Citation2008) which is not well known. Moreover, the taxonomy of this species is controversial (Smith & Xie Citation2008). In genus Petaurista, only two complete mitochondrial genomes have been reported: 16 507 bp Petaurista alborufus JQ743657 and 16502 bp Petaurista hainana JX572159 (Kong et al. Citation2015). Given this, it seems particularly important to obtain the mitochondrial genome sequence of this species. We expect that this work can provide some available information for clarifying the taxonomic dispute surrounding P. elegans caniceps.
In this study, the P. elegans caniceps was obtained from wild field in Yunnan, China (Geospatial coordinates: 27°35’447″ °N, 99°18’068″ °E, with accession No. PE-15-01, China West Normal University). We obtained the complete mitochondrial genome of P. elegans caniceps, and submitted it to the GeneBank database with an accession number KU579289. The complete genome is 16 520 bp in size and contains 22 tRNA genes, 13 protein-coding genes (PCGs), 12S rRNA, 16S rRNA and 1 control region. The base composition is: A, 32.45%; T, 31.75%; C, 23.47%; and G, 12.33%. All of the PCGs are in the heavy chain except ND6. ATG is the start codon for all protein-coding genes. Five genes end with an incomplete stop codon TA– (ND2, ND3, and ND5) or T–– (COX3 and ND4). ND6 and Cytb genes terminate with TAG and AGA, respectively, and the other six genes end on the usual TAA stop codons.
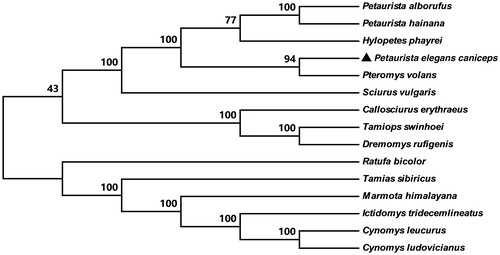
The mitochondrial genome has been broadly applied in population genetics and phylogenetic (Singh Citation2008; Galtier et al. Citation2009). To further validate the taxonomy and phylogenetic relationships, phylogenetic analysis was constructed by whole mitogenome sequences with 14 closely related taxa. A neighbour-joining (NJ) tree was constructed using MEGA 6 (Tamura et al. Citation2013) with 1000 bootstrap replicates (). From the phylogenetic tree, we can see that P. elegans caniceps first clustered with Pteromys volans, and then together with other four species, including two Petaurista sp. (Petaurista alborufus and Petaurista hainana), Hylopetes phayrei and Sciurus vulgaris forming a single branch.
Figure 1. NJ tree inferred from the nucleotide sequence of whole mitogenome. The numbers above branches specify bootsrap percentages (1000 replicates). The species were selected from Petaurista elegans caniceps; Pteromys volans (JQ230001); Hylopetes phayrei (KC447305); Petaurista alborufus (JQ743657); Petaurista hainana (JX572159); Sciurus vulgaris (AJ238588); Dremomys rufigenis (KC447304); Tamiops swinhoei (KP027416); Tamias sibiricus (KF668525); Callosciurus erythraeus (KM502568); Ictidomys tridecemlineatus (KP698974); Cynomys leucurus (KP326309); Cynomys ludovicianus (KP326310); Marmota himalayana (JX069958); Ratufa bicolor (KF575124).
Acknowledgements
We are grateful to our field assistants Shuikai Feng. We thank Baimaxueshan Nature Reserve for our work permit. We greatly appreciate Dr. Ali Krzton for revising English language for this manuscript. We thank Foundation of key Laboratory of Southwest China Wildlife Resources Conservation (Ministry of Education), China West Normal University.
Disclosure statement
The authors have declared no conflicting interests. The authors alone are responsible for doing the research and writing the paper.
References
- Galtier N, Nabholz B, Glémin S, Hurst GDD. 2009. Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Mol Ecol. 18:4541–4450.
- Kong L, Wang Y, Cong H, Wang W, Li Y. 2015 Characterization of the complete mitochondrial genome of the Hainan giant flying squirrel Petaurista hainana (Rodentia: Sciuridae). Mitochondrial DNA. 26: 871–872.
- Singh TR. 2008. Mitochondrial gene rearrangements: new paradigm in the evolutionary biology and systematics. Bioinformationn 3:95–7.
- Smith AT, Xie Y. 2008. A guide to the mammals of China. Vol. Xiii. Princeton (NJ): Princeton University Press.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30:2725–2729.
