Abstract
Panicum miliaceum is the most important and ancient domesticated crops in the world. The complete chloroplast genome of P. miliaceum was sequenced using the Illumina Hiseq platform (Illumina Inc., San Diego, CA). The chloroplast genome of P. miliaceum was 139,929 bp in length, with 38.60% GC content. It contains a pair of inverted repeats (IRs) (22,723 bp) which were separated by a large single copy (LSC) (81,918 bp) and a small single copy region (SSC) (12,565 bp). A total of 132 genes were annotated, which included 84 protein coding genes, 40 tRNA genes and eight rRNA genes. The neighbour-joining (NJ) phylogenetic analysis with the reported chloroplast genomes showed that P. miliaceum chloroplasts are most closely related to those of the Gramineae family.
Broomcorn millet (Panicum miliaceum) was one of the most important and ancient domesticated crops in the world (Lu et al. Citation2009). They were staple foods in the semiarid regions of East Asia (China, Japan, Russia, India and Korea) and even in the entire Eurasian continent before the popularity of rice and wheat (Fuller Citation2006), and are still important foods in these regions today (Li & Wu Citation1996; Lu et al. Citation2009).
Chloroplasts play an important role in photosynthesis in green plants (Neuhaus & Emes Citation2000). The plant chloroplast genome consists of two large inverted repeats separated by a large single copy (LSC) region and a small single copy (SSC) region (Palmer Citation1985). The DNA sequence of the chloroplast genome can be used as a super barcode or a resource for research in phylogeograhy, genetic diversity and evolution. Although many members of the Gramineae are economically important and had complete chloroplast genome data available on pubic database on website, the common millet (P. miliaceum) has no its complete chloroplast sequence published.
For this study, we collected the fresh, healthy leaves from P. miliaceum growing in the site (39°08'N, 111°14'E), which is located near the junction between the Loess Plateau and the North China Plain at an elevation of 1086 m. DNA was extracted using CTAB-based protocol described by Gawel (Citation1991). We sequenced the complete chloroplast genome of P. miliaceum with the Illumina HiSeq sequencing platform (Novogene, Beijing, China). We assembled the chloroplast genome using Mira and annotated with software CpGAVAS (Liu et al. Citation2012). We determined the complete chloroplast genome sequence of P. miliaceum was 139,929 bp in length. The GC content was 38.6%. Chloroplasts are typically AT rich, and the GC content of the P. miliaceum chloroplast was similar to values previously reported for most other Gramineae species (e.g. 39.0% in Chikusichloa aquatica; Zhang et al. Citation2015). The LSC and SSC contained 81,918 bp and 12,565 bp, respectively, while the IR was 22,723 bp in length. This chloroplast genome contained 132 functional genes, including 84 protein-coding genes, 30 tRNA genes and eight rRNA genes. There were 14 protein-coding genes, 16tRNA and all eight rRNA genes duplicated in the IR regions. The LSC region contained 60 protein-coding and 23 tRNA genes, whereas the SSC region contained 10 protein-coding and 1tRNA genes. Fourteen genes contained one or two introns, including the protein-coding genes, rps16, atpF, ycf3, petB, petD, rpl16, rpl2, ndhB, ndhA and rps12.
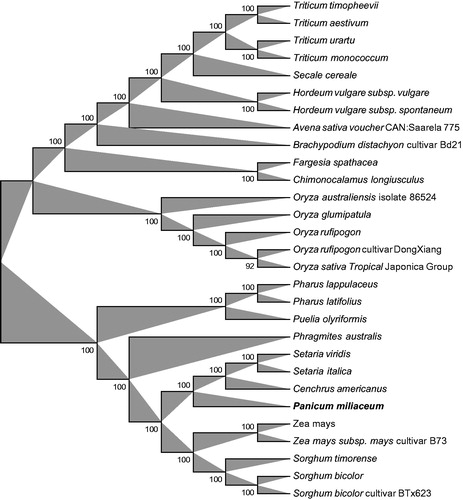
The phylogenetic relationship of P. miliaceum to other plant families was evaluated by comparing the 29 species of Gramineae chloroplast. A neighbour-joining (NJ) analysis was performed using the software Geneious 8.0.2 (http://www.geneious.com/) (). The phylogenetic tree showed that P. miliaceum (Gramineae) was most closely related to members of genus Cenchrus and the genus Setaria (). The newly characterized P. miliaceum complete chloroplast genome will provide essential data for further study on the phylogeny and evolution of the genus Panicum and of the Gramineae, for molecular breeding, and potentially for genetic engineering.
Figure 1. Phylogenetic relationships among 28 whole chloroplast genomes of Gramineae family. The 29 species can be divided into two independent clades. Bootstrap support values are given at the nodes. Chloroplast genome accession number used in this phylogeny analysis: Triticum timopheevii: NC024764; Triticum aestivum: KC912694; Triticum urartu: NC021762; Triticum monococcum: NC021760; Secale cereale: NC021761; Hordeum vulgare subsp. Vulgare: KC912689; Hordeum vulgare subsp. spontaneum: KC912688; Avena sativa voucher CAN:Saarela 775: NC027468; Brachypodium distachyon cultivar Bd21: EU325680; Fargesia spathacea: NC024716; Chimonocalamus longiusculus: NC024714; Oryza australiensis isolate 86524: NC024608; Oryza glumipatula: KM881640; Oryza rufipogon: NC017835; Oryza rufipogon cultivar DongXiang: KF562709; Oryza sativa Tropical Japonica Group: KT289404; Pharus lappulaceus: NC023245; Pharus latifolius: NC021372; Puelia olyriformis: NC023449; Phragmites australis: NC022958; Setaria viridis: NC028075; Setaria italica: KJ001642; Cenchrus americanus: KJ490012; Zea mays: NC001666; Zea mays subsp. mays cultivar B73: KF241981; Sorghum timorense: NC023800; Sorghum bicolor: NC008602; Sorghum bicolor cultivar BTx623: EF115542. Bold and italic were the sample as Panicum miliaceum in this study.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Funding
This work was supported by the Doctoral Scientific Fund Project of Shanxi Academy of Agricultural Sciences (No. YBSJJ1410) and the Earmarked Fund for China Agriculture Research System (Nos CARS-07-13.5).
References
- Fuller DQ. 2006. Agricultural origins and frontiers in South Asia: a working synthesis. J World Prehist. 20:1–86.
- Gawel RLJ. 1991. A modified CTAB DNA extraction procedure for musa and ipomoea. Plant Mol Biol Rep. 9:262–266
- Li Y, Wu SZ. 1996. Traditional maintenance and multiplication of foxtail millet (Setaria italica. (L.) P. Beauv.) landraces in China. Euphytica. 87:33–38.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715.
- Lu H, Zhang J, Liu K, Wu N, Li Y, Zhou K, Yed M, Zhang T, Zhang H, Yang X, et al. 2009. Earliest domestication of common millet (Panicum miliaceum) in East Asia extended to 10,000 years ago. Proc Natl Acad Sci USA. 106:7367–7372.
- Neuhaus HE, Emes MJ. 2000. Nonphotosynthetic metabolism in plastids. Annu Rev Plant Physiol Plant Mol Biol. 51:111–140.
- Palmer JD. 1985. Comparative organization of chloroplast genomes. Annu Rev Genet. 19:325–354.
- Zhang J, Zhang D, Shi C, Gao J, Gao LZ. 2015. The complete chloroplast genome sequence of Chikusichloa aquatica (Poaceae: Oryzeae). Mitochondrial DNA. [Epub ahead of print]. DOI:10.3109/19401736.2015.1053058
