Abstract
Artemisia argyi, called wormwood, is widely distributed in northeastern Asia. The complete chloroplast genome sequence of A. argyi was generated by de novo assembly using whole genome next generation sequences. The complete chloroplast genome sequence of A. argyi is 151 192 bp in size. It is composed of a large single-copy (LSC), a small single-copy (SSC) and two inverted repeat (IR) regions of 82 930 bp, 18 344 bp and 24 959 bp, respectively. Overall GC contents of the genome were 37.46%. The A. argyi chloroplast genome has a total of 114 genes including 80 protein-coding genes, 30 tRNA genes and four rRNA genes. Phylogenetic analysis based on the chloroplast genome demonstrated that A. argyi is most closely related to Artemisia montana.
Artemisia argyi is a perennial herb as a member of the Artemisia belonged to the family Asteraceae (Bremer Citation1994) and widely distributed in Asia, such as China, Mongolia, Japan and Korea. Artemisia species are naturalized in dry and semi-arid habitats and mentioned as “wormwood”. Artemisia argyi, referred to as “Aeyup” in Korea, has been used for the treatment of colic pain, vomiting, and irregular uterine bleeding (Zhao et al. Citation1994). Despite its useful applications, there has been little report of molecular and genomic resources (Vallès & McArthur Citation2001; Shoemaker et al. Citation2005; Liu et al. Citation2013). In this article, the chloroplast genome sequence of medicinal plant, A. argyi, was completely characterized.
The plant samples of A. argyi were collected from Eumseong (Latitude: 36° 56′38.68″N, Longitude: 127° 45′17.60″E), Korea and identified by Dr. JH Lee, Department of Herbal Crop Research, NIHHS, RDA. A voucher specimen (MPS003336) is deposited at Korea Medicinal Resources Herbarium, Eumseong Korea. Whole genome sequencing was performed using Illumina genome analyzer (Hiseq1000, Illumina, San Diego, CA) platform at the in-house facility (Genomics Division, NAAS, RDA, Jeonju, Korea). De novo assembly was performed using CLC genome assembler (v. beta 4.6, CLC Inc., Aarhus, Denmark), as mentioned in Kim et al. (Citation2015). We obtained three main contigs from de novo genome assembly. Main contigs cover the entire chloroplast genome of reported A. frigida chloroplast sequence (Liu et al. Citation2013). Three contigs could be joined as one single circular complete sequence by manual editing. Gene annotation was conducted using DOGMA (Wyman et al. Citation2004) and manual curation through comparison with published chloroplast genomes deposited on NCBI. Inverted repeats were identified using Tandem repeat finder (https://tandem.bu.edu/trf/trf.html). Simple sequence repeat (SSR) motifs were identified using NWISRL (http://ssr.nwisrl.ars.usda.gov/stop2.php). The complete chloroplast genome of A. argyi was submitted to GenBank under the accession no. KM386991.
The complete chloroplast genome of A. argyi is 151,192 bp in size, which is composed of a large single-copy (LSC), a small single-copy (SSC) and two inverted repeat (IR) regions of 82 930 bp, 18 344 bp and 24 959 bp, respectively. A. argyi chloroplast genome has a GC content of 37.46%. It contains 80 protein-coding genes, 30 tRNA genes and four rRNA genes. According to comparative analysis among three Artemisia species, sequence identities of whole chloroplast genomes were revealed like 99.8% between A. argyi and A. montana, otherwise, 99.3% between A. argyi and A. frigida. A. argyi was more closely related with A. montana than A. frigida according to analysis based on chloroplast genome. Simple sequence repeats (SSRs) of chloroplast genome sequence were found 92 in A.argyi, most of them were tri-nucleotide SSR motifs and commonly distributed throughout the genome.
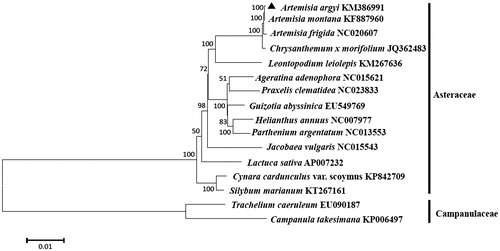
The phylogeny of A. argyi to other species in the Asteraceae (family Compositae) was generated. A phylogenetic tree of A. argyi was constructed using entire chloroplast protein-coding sequences of A. argyi with 14 published species in family Asteraceae by a maximum likelihood (ML) analysis of MeGA 6.0 (MEGA Inc., Englewood, NJ) (Tamura et al. Citation2013). The phylogenetic tree indicated that A. argyi was grouped to the genus Artemisia, as expected and most closely related to A. montana in the genus Artemisia ().
Figure 1. ML phylogenetic tree of Artemisia argyi with related 14 species of the family Asteraceae based on entire chloroplast protein-coding genes using Mega 6.0 with ML method. Numbers at the nodes are bootstrap values from 1000 replicates. Trachelium caeruleum (EU090187) and Campanula takesimana (KP006497) in Campanulaceae were set as the outgroup.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Funding information
This work was carried out with the support of National Academy of Agricultural Science [Project no. PJ010889] and Cooperative Research Program for Agriculture Science and Technology Development [Project title: National Agricultural Genome Program, Project no. PJ010457], Rural Development Administration, Republic of Korea.
References
- Bremer K. 1994. Asteraceae: cladistics and classification. Portland (OR): Timber Press.
- Zhao AC, Kiyohara H, Yamada H. 1994. Anti-complementary neutral polysaccharides from leaves of Artemisia princeps. Phytochemistry. 35:73–77.
- Kim K, Lee SC, Lee J, Lee HO, Joh HJ, Kim NH, Park HS, Yang TJ. 2015. Comprehensive survey of genetic diversity in chloroplast genomes and 45S nrDNAs within Panax ginseng species. PLoS One. 10:e0117159.
- Liu Y, Huo N, Dong L, Wang Y, Zhang S, Young HA, Feng X, Gu YQ. 2013. Complete chloroplast genome sequences of Mongolia medicine Artemisia frigida and phylogenetic relationships with other plants. PLoS One. 8:e57533.
- Shoemaker M, Hamilton B, Dairkee SH, Cohen I, Campbell MJ. 2005. In vitro anticancer activity of twelve Chinese medicinal herbs. Phytother Res. 19:649–651.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Vallès J, McArthur ED. 2001. Artemisia systematics and phylogeny: cytogenetic and molecular insights. Shrubland Ecosyst Genet Biodivers. 21:67–74.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
