Abstract
The badger is a common carnivorous animal maintain ecosystem by regulating the population of their prey. However, genomic information such as sequence polymorphisms has been restricted until recently. In this study, we have sequenced and assembled complete mitogenomes of Asian badgers, Meles leucurus amurensis, and polymorphic sites were identified. A total 182 singleton polymorphic sites were identified, and the 92 sites were located in protein-coding genes. Phylogenetic analysis showed that Asian badgers are evolutionary closed to Japanese badgers rather than European badgers. This study will provide important genomic information to assign taxon of species and to identify species of mustelids.
Meles leucurus amurensis is one of the Asian badger subspecies, which is widely distributed from the East China to Korean Peninsula. These small carnivorous animals are important predators regulating population of their prey in forest, and maintaining ecosystem of their habitats (Lindström Citation1989).
A few studies have tried to classify badgers in genus Meles. It has been considered first that three species compose the genus (Abramov & Puzachenko Citation2006). However, phylogenetic analysis of Eurasian badgers suggested that the genus would be classified into four groups (Marmi et al. Citation2006) including badgers from Southwest Asia, M. canescens, showing distinctive phylogenetic relationships with other Eurasian badgers (Del Cerro et al. Citation2010). Considering the M. canescens was first classified as a subspecies of European badgers, M. meles (Blanford Citation1875), reconsidering the classification of badgers in Meles is required. However, genomic information of Eurasian badgers such as polymorphisms, which are important information for tracing divergence of species, is limited. In this study, complete mitogenomes of the two Asian badgers, M. leucurus amurensis, were determined (GenBank accession no. KU707935-6) and compared with the reported Asian badger mitogenome (Zhou et al. Citation2015) to observe the mitogenomic polymorphisms in M. leucurus amurensis. RNA was extracted from kidney, liver, and brain of slaughtered individuals (BioSample IDs: SAMN04508153-8) donated from a local farm (35.0554N; 126.3342E), and assessed to RNA-seq following by assembled by alignment to previously reported mitogenome sequences. Mitogenome annotation was conducted by DOGMA (Wyman et al. Citation2004) and polymorphic sites were searched using DNaspV5 (Librado & Rozas Citation2009). Phylogenetic tree was constructed with complete mitogenome sequences of mustelids using MEGA6 under built-in Tamura–Nei and maximum-likelihood model with 1000 bootstrap replication (Tamura et al. Citation2013).
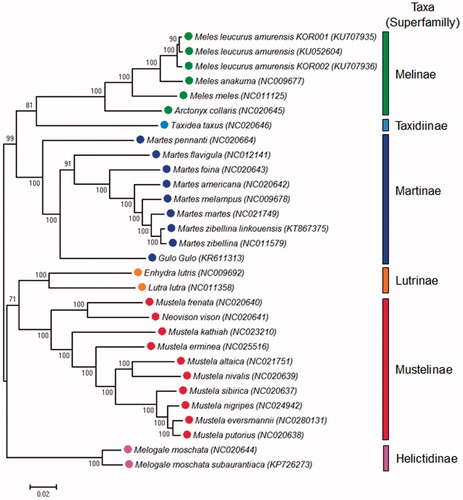
The complete mitogenome sequences of the individuals were 16496–7 bp lengths, and AT/GC contents were about 61.1% and 38.9%. Three and 179 sites were indels and singleton polymorphic sites (1.1% of the mitogenome sequences), respectively. Nine and 89 sites were non-synonymous and synonymous polymorphic sites (Supplementary Table 1). Phylogenetic tree showed that two Asian badgers in Korea showed closer pairwise distance each other compared to Asian badgers in China (). This would mean the badgers in Korean Peninsula might have specific mitogenomic characters distinguished from the Asian badgers in China. In addition, Asian badgers showed closer evolutionary distances with Japanese badgers rather than European badgers despite the geographical seclude by the East Sea (). And organisms in Melinae including Asian badger have the closer evolutionary distances with Martinae and Taxidiinae species compared with mustelids in other superfamily. Identified polymorphisms and enlarged mitogenomic information of Asian badgers in this study could be used to understand speciation and support establishment of taxa in mustelids.
Disclosure statement
The authors report that they have no conflicts of interest.
Funding information
This study was supported by the 2015 Research Grant from Kangwon National University (No. 520150139), Republic of Korea.
Supplementary_Table_1._Polymorphic_sites.docx
Download ()References
- Abramov A, Puzachenko AY. 2006. Geographical variability of skull and taxonomy of Eurasian badgers (mustelidae, meles). Zoologicheskii Zhurnal. 85:641–655.
- Blanford W. 1875. Xl. – descriptions of new mammalia from persia and balúchistan. Ann Mag Nat Hist. 16:309–313.
- Del Cerro I, Marmi J, Ferrando A, Chashchin P, Taberlet P, Bosch M. 2010. Nuclear and mitochondrial phylogenies provide evidence for four species of Eurasian badgers (carnivora). Zool Scr. 39:415–425.
- Librado P, Rozas J. 2009. Dnasp v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 25:1451–1452.
- Lindström E. 1989. The role of medium-sized carnivores in the Nordic boreal forest. Finnish Game Res. 46:53–63.
- Marmi J, Lopez-Giraldez F, Macdonald DW, Calafell F, Zholnerovskaya E, Domingo-Roura X. 2006. Mitochondrial DNA reveals a strong phylogeographic structure in the badger across Eurasia. Mol Ecol. 15:1007–1020.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. Mega6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with dogma. Bioinformatics. 20:3252–3255.
- Zhou W, Yu L, Tan B, Liu Y, Zhang L, Hua Y. 2015. Phylogenetic relationship of Asian badger meles leucurus amurensis revealed by complete mitochondrial genome. Mitochondrial DNA. [Epub ahead of print]. DOI: 10.3109/19401736.2015.1127365.