Abstract
In this study, complete nucleotide sequences of the mitochondrial genome of a nudibranch species, Hypselodoris festiva (A. Adams, 1861) were determined and characterized. The mitogenome size is 14 880 bp. This is the longest among the known nudibranch mitochondrial genomes. Furthermore, phylogenetic relationship of H. festiva in the Nudibranchia reconstructed due to amino acid sequences of mitochondrial protein coding genes. This is the sixth record for complete mitochondrial genome of the Nudibranchia.
Hypselodoris festiva (A. Adams, 1861) is a common nudibranch species which lives in the intertidal areas of Korea and Japan (Choe Citation1992; Okutani Citation2000). There is no known complete mitochondrial sequence information for the species in the nucleotide databases even though molecular phylogenetic study for species of the Chromodorididae was accomplished (Johnson & Gosliner Citation2012). In this study, H. festiva was collected from a subtidal area of Seongsan (33°22′24″N, 126°54′5″E), Jeju island, Korea by scuba diving (Marine Biodiversity Institute of Korea Accession number MABIK MO00157620). The complete mitochondrial genome of H. festiva was characterized in the study and furthermore, by using amino acid sequences of protein coding genes in the mitochondrial genome phylogenetic relationships of H. festiva in the Nudibaranchia was investigated.
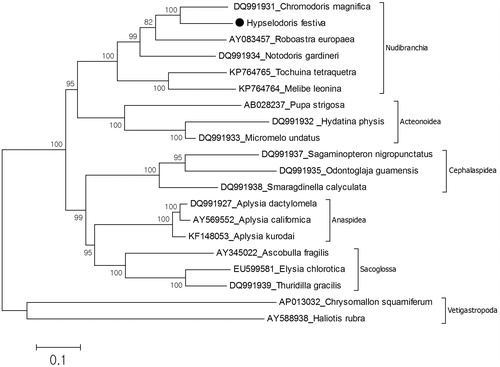
The size of mitochondrial genome of H. festiva is 14,880 bp (GenBank accession no. KU365323). This is the sixth record for complete mitochondrial genome of the Nudibranchia and the longest among the known nudibranch mitochondrial genomes. The nucleotide composition of the genome is 31.1% A, 14.3% C, 16.7% G, 37.9% T. AT content (69%) of the mitochondrial genome is also the highest among nudibranch mitochondrial genomes. The mitochondrial genome is composed of 13 protein coding, two ribosomal RNA and 22 tRNA genes. The gene order of the mitochondrial genome is almost the same to the other nudibranch mitochondrial genomes, while the position of tRNA-Ser and ND4 gene moved to between tRNA-Cys and tRNA-Gln. The change of genomic position is confirmed by PCR and resequencing. There are five overlapping regions ranging from 1 to 20 bp. The mitochondrial genome has 26 intergenic sequences, showing the longest one positioned between ND4 and tRNA-Gln. The phylogenetic tree () suggests that H. festiva is placed in the monophyletic Nudibranchia and Chromodoris magnifica is the closest species to H. festiva. Due to our results the Nudibaranchia has sister group relationship with Acteonidea. This data supports the result of previous study based on mitochondrial genome (Medina et al. Citation2011). The present study provides additional data for Nudibranchia phylogeny.
Figure 1. Phylogenetic relationship of the Hypselodoris festiva. The complete mitochondrial genomes of the gastropod species were retrieved from the GenBank for the analysis of phylogenetic relationship of H. festiva. Phylogenetic tree based on 5019 concatenated amino acid sequences from 12 of the 13 protein-coding mitochondrial genes (ATP8 gene was excluded from the analysis) was reconstructed by MEGA (Kumar et al. Citation1993) with maximum-likelihood analyses based on mtREV with Freqs (+F) model. Bootstrap method used 1000 replicates to know statistical support. Two species of the Vetigastropoda were used as an outgroup. The specimen of H. festiva was preserved in 97% ethanol and DNA was extracted from foot. Paired end reads generated from a mitochondrial enriched genomic library. The reads were assembled and annotated by using MITObim (Hahn et al. Citation2013) and MITOS (Bernt et al. Citation2013), respectively.
Funding information
This work was supported by National Marine Biodiversity Institute under Research Program (Number 2015M00300).
Disclosure statement
The authors report no conflicts of interest.
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Choe BL. 1992. Illustrated encyclopedia of fauna and flora of Korea vol. 33, Mollusca (II). Seoul, Korea: Ministry of Education (in Korean).
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Johnson RF, Gosliner TM. 2012. Traditional taxonomic groupings mask evolutionary history: a molecular phylogeny and new classification of the chromodorid nudibranchs. PLoS One 7:e33479. doi:10.1371/journal.pone.0033479.
- Kumar S, Tamura K, Nei M. 1993. MEGA: molecular evolutionary genetics analysis. University Park (PA): The Pensilvania State Univ.
- Medina M, Lal S, Vallès Y, Takaoka TL, Dayrat BA, Boore JL, Gosliner T. 2011. Crawling through time: transition of snails to slugs dating back to the Paleozoic, based on mitochondrial phylogenomics. Mar Genom. 4:51–59.
- Okutani T. 2000. Marine Mollusks in Japan. Tokyo, Japan: University of Tokyo University Press.
