Abstract
In this study, the complete mitochondrial genome of the Bowmouth guitarfish Rhina ancylostoma (Rajiformes, Rhinobatidae) was first determined. The total length of this circle DNA was 17 217 bp, consisted of 37 genes with a typical gene order in vertebrate mitogenome. It had 42 bp short intergenic spaces and 40 bp overlaps. The nucleotide composition in R. ancylostoma was as follows: A, 33.0%; C, 25.1%; G, 12.6% and T, 29.3%. Two start codons (GTG and ATG) and two stop codons (TAG and TAA/T) were used in the protein-coding genes. The 22 tRNA genes were ranged from 67 bp (tRNA-Ser2) to 75 bp (tRNA-Leu1). The phylogenetic result showed that R. ancylostoma was clustered with the Rhinobatos.
Keywords:
The Bowmouth guitarfish Rhina ancylostoma (Rajiformes, Rhinobatidae), a widely distributed Indo-west Pacific inshore species, mainly occurs on or close to the seabed, which was a viviparous mammals (Carpenter et al. Citation1997; Compagno & Last Citation1999). It was very little known about the molecular and genetic research of R. ancylostoma. In this study, we first determined the complete mitochondrial genome of R. ancylostoma and analyzed the phylogenetic relationship in Rajiformes.
One specimen of R. ancylostoma (Museum of marine biology of Wenzhou Medical University, voucher XM2012032117) captured in the Taiwan Strait, was collected from a fish market in Xiamen, China. The experimental protocol and data analysis methods followed Chen et al. (Citation2013). Including R. ancylostoma, 13 species of Rajiformes, with the complete mitogenomes available in the Genbank, were selected to construct the phylogenetic tree using the Bayesian method. The outgroup was Dasyatis akajei (Myliobatoformes).
The complete mitochondrial DNA of R. ancylostoma was 17 217 bp (Genbank accession no. KU721837), with a typical mitogenomic organization and gene order as most vertebrates, containing 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes and 1 non-coding control region. The nucleotide composition in R. ancylostoma was as follows: A, 33.0%; C, 25.1%; G, 12.6% and T, 29.3%. The whole mitogenome had 42 bp short intergenic spaces located in 15 gene junctions and 40 bp overlaps located in seven gene junctions. Two start codons (GTG and ATG) and two stop codons (TAG and TAA/T) were used in the protein-coding genes, and most of them shared common initial codon ATG and terminal codon TAA/T. A non-standard initial codon GTG was owned by the COI gene, which was common in vertebrates (Slack et al. Citation2003). The COII and ND4 genes were terminated with a single T, which could be extended to complete TAA through polyadenylation after transcriptions (Ojala et al. Citation1981). The 22 tRNA genes interspersed between the rRNAs and protein-coding genes, ranging from 67 bp (tRNA-Ser2) to 75 bp (tRNA-Leu1), while the 12S and 16S rRNA genes were located between the tRNA-Phe and tRNA-Leu1 genes, separated by the tRNA-Val genes. A 39 bp inserted sequence was identified as the origin of light-strand replication (OL) between tRNA-Asn and tRNA-Cys genes with a stem-loop structure. The control region was 1499 bp, presenting a high A + T content (68.0%). There were a 48 bp motif repeated five times and a 46 bp motif repeated seven times in the control region. The high similarity (71.43%) suggested that they were homologous.
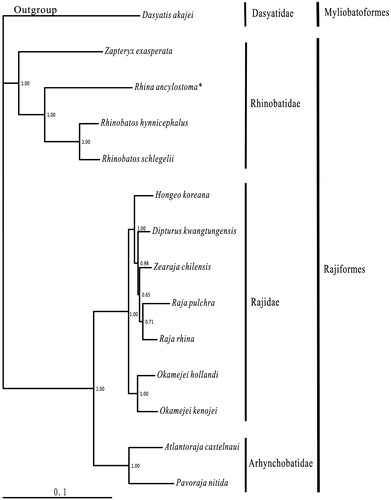
Within the Rajiformes, 13 available species belonged to three families with the (Rhinobatidae + (Rajidae + Arhynchobatidae)) relationship were consistent to the morphological result (McEachran & Dunn Citation1998). Rhina ancylostoma was sister to the Rhinobatos clade, then this clade clustered to Zapteryx exasperata ().
Figure 1. Phylogenetic position of Rhina ancylostoma Dasyatis akajei (NC_021132.1) was selected as the outgroup. The 13 species of Rajiformes were Zapteryx exasperata (NC_024937.1), Rhina ancylostoma (KU721837), Rhinobatos hynnicephalus (NC_022841.1), R. schlegelii (NC_023951.1), Hongeo koreana (NC_021963.1), Dipturus kwangtungensis (NC_023505.2), Zearaja chilensis (KJ913073.2), Raja pulchra (NC_025498.1), Raja rhina (KC914434.1), Okamejei hollandi (KP756687.1), O. kenojei (NC_007173.1), Atlantoraja castelnaui (NC_025942.1), and Pavoraja nitida (NC_024599.1).
Disclosure statement
This study was supported by Ministry of Science and Technology of Zhejiang Province (2013F50015). The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Carpenter KE, Krupp F, Jones DA, Zajonz U. 1997. Living marine resources of Kuwait, eastern Saudi Arabia, Bahrain, Qatar, and the United Arab Emirates. FAO species identification field guide for fishery purposes. Rome: FAO. p. 293.
- Chen X, Ai WM, Ye L, Wang XH, Lin CW, Yang SY. 2013. The complete mitochondrial genome of the grey bamboo shark (Chiloscylliumgriseum) (Orectolobiformes: Hemiscylliidae): genome characterization and phylogenetic application. Acta Oceanol Sin. 32:59–65.
- Compagno LJV, Last PR. 1999. Rhinidae (=Rhynchobatidae). Wedgefishes. In: Carpenter KE, Niem V, editors. FAO identification guide for fishery purposes. The Living Marine Resources of the Western Central Pacific. Rome: FAO. p. 1418–1422.
- Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature. 290:470–474.
- McEachran JD, Dunn KA. 1998. Phylogenetic analysis of skates, a morphologically conservative clade of elasmobranchs (Chondrichthyes: Rajidae). Copeia. 1998:271–290.
- Slack KE, Janke A, Penny D, Arnason U. 2003. Two new avian mitochondrial genomes (penguin and goose) and a summary of bird and reptile mitogenomic features. Gene. 302:43–52.
