Abstract
The complete mitochondrial genome sequences of Pleurobranchaea novaezealandiae and Pleurobranchaea sp. are first described and analyzed in this study. It is 14,531 bp and 14,709 bp in length, respectively. The base composition of the genome with A + T bias are 66.41% and 68.36%. There are 29 noncoding regions found throughout the mitogenome of P. novaezealandiae and 30 noncoding in Pleurobranchaea sp., ranging in size from 2 to 294 bp. The phylogenetic tree based on 10 mitogenome, including 1 prosobranchia, 6 opisthobranchia and 3 pulmonata was analyzed in the paper. The results showed that the opisthobranchia and pulmonata were clustered respectively, and the P. novaezealandiae and Pleurobranchaea sp. were the closest to the Aplysia californica in our analysis.
Pleurobranchaea novaezealandiae (voucher number: ASTM-Mo-P541) belongs to Mollusc, Gastropoda, Opisthobranchia, Pleurobranchidae, which has a widespread domestic distribution, such as Bohai Sea, Yellow Sea, Paracel Islands and HongKong, and Japan is its main foreign inhabitancy. Pleurobranchaea novaezealandiae is characterized by small body. It presents oblong shape, with a bulge on its back and irregular mastoid processes on its surface. The front of its head extends bilaterally to form flat shape, with small serration at its edge and two pairs of antennae. It has large ovate-oblong feet, which is exposed outside the margin of mantle. It has a blue-yellow back, while its ventral side appears dark maroon.
Pleurobranchaea sp. (voucher number: ASTM-Mo-P547) belongs to Mollusc, Gastropoda, Opisthobranchia, Pleurobranchidae, a small creature inhabiting at shallow. It has oval shape, sharp tail and resembles slug. Besides, with a bulge on its back, its surface emerges white mastoid processes. Pleurobranchaea sp. is bilaterally symmetric taking pleopod as axle wire, and gills located at the front of head can assist to breathe, together with the secondary gills beneath mantle and the bilateral gills of pleopod.
The complete mitochondrial genome of P. novaezealandiae and Pleurobranchaea sp. were sequenced and characterized in this article, and the samples of P. novaezealandiae and Pleurobranchaea sp. were collected from Ganyu, Jiangshu Province, China. Before further processing, specimens were stored in ultra-low temperature freezer.
The complete mitochondrial genome of P. novaezealandiae and Pleurobranchaea sp. were 14,531 bp and 14,709 bp in length, respectively, and have been deposited in GenBank with accession No. KU365727 and No. KU365728. They consist of 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes as shown in and . The ATG, ATT and TTG are used as the start codons, which are very common in invertebrates (Grande et al. Citation2008), but only ATP8 starts with TTG. Except for COX3 with an incomplete stop codon “T––”, the remaining protein-coding genes stop with the TAG or TAA. Using the tRNA scan-SE 1.21 (Lowe & Eddy Citation1997), 22 tRNA genes were found to fold into a typical cloverleaf secondary structure. The overall basic composition of the heavy strand in P. novaezealandiae is 36.53% A, 29.88%T, 19.67% G and 13.92% C, with an AT content of 66.41%. Similarly, the overall base composition of the heavy strand in Pleurobranchaea sp. is 40.02% A, 28.34%T, 18.46% G and 13.18% C, with an AT content of 68.36%. The AT content is higher than the content of GC, as generally shown in bivalvia mitochondrial genomes (Wang et al. Citation2010).
Table 1. Mitochondrial genome of the Pleurobranchaea novaezealandiae.
Table 2 Mitochondrial genome of the Pleurobranchaea sp.
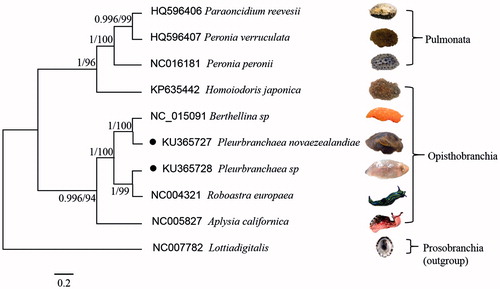
The phylogenetic tree () was generated using the MrBayes soft analyses and Maximum Likelihood method (Tamura et al. Citation2011) from amino acid composition of complete mitochondrial genomes. The results showed that the opisthobranchia and pulmonata were clustered respectively, and the P. novaezealandiae and Pleurobranchaea sp. were the closest to the Aplysia californica in our analysis. It constituted a framework for phylogeny evolution analysis, systematic classification of other euthyneurans (pulmonates and opisthobranchs) (Liu et al. Citation2015). The comparative mitogenomic analysis of gastropods may provide valuable phylogenetic information.
Funding information
The study was supported by the National Natural Science Foundation of China (No.41276157) and Shanghai Universities First-class Disciplines Project of Fisheries.
Acknowledgements
We appreciate anonymous reviewers for providing valuable comments on this article.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
References
- Grande C, Templado J, Zardoya R. 2008. Evolution of gastropod mitochondrial genome arrangements. BMC Evol Biol. 8:1–15.
- Liu C, Wu X, Shen HD. 2015. Complete mitochondrial genome of Vaginulus alte and Homoiodoris japonica. Mitochondrial DNA. Online 5 August 2015. DOI:10.3109/19401736.2015.1066345.
- Lowe TM, Eddy SR. 1997. TRNAscan-SE 1.21; [cited 2011 Apr 5]. Available from: http://lowelab. ucsc. edu/tRNA scan-SE/.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739.
- Wang HX, Zhang SP, Li Y, Liu BZ. 2010. Complete mtDNA of Meretrix lusoria (Bivalvia: Veneridae) reveals the presence of an atp8 gene, length variation and heteroplasmy in the control region. Comp Biochem Physiol Part D. 5:256–264.