Abstract
In this paper, we determined the complete mitochondrial genome of Chinese Daphnia carinata for the first time by the long and accurate polymerase chain reaction and primer-walking methods. It was 15,245 bp in length, with an A + T content of 70.35%, containing 37 typical animal mitochondrial genes and an A + T-rich region. The COI gene started with ACTA. All the 22 typical tRNA genes had a classical cloverleaf structure except for trnS1, in which the D-stem pairings in the DHU arm were absent.
Water fleas are an important component of the microcrustacean zooplankton, their habitats are mostly continental fresh and saline waters (Forró et al. Citation2008). Daphnia carinata has become a well-known model species in studying evolutionary biology, environmental biology and ecology (Miner et al. Citation2013). Only the complete mitochondrial DNA of North American Daphnia pulex (GenBank accession no. AF117817) and the Chinese Daphnia pulex (GenBank accession no. KT003819) and the Chinese Daphnia magna (GenBank accession no. KP296147) have been released in Genbank database. And other partial mitochondrial genome DNA of Daphnia has also been published; however, this is far from enough. Gu et al. (Citation2013) had named Daphnia similoides sinensis for this D. carinata, by comparing the sequences of COI with the similarity of 99%. So the standard of morphology species identification could not meet taxonomy studies. It is, thus, worthy to do the further researches for molecular biology of Daphnia so far, advancing our understanding of Cladoceran diversity and evolution.
Using the long and accurate polymerase chain reaction and primer-walking methods, we sequenced and characterized the complete mitochondrial genome of Chinese D. carinata (sampled from Chaohu, Anhui province in China) for the first time (GenBank accession no. KP721459). The total length is 15,245 bp with an A + T content of 70.35%. provides detailed information about the mitogenome. The D. carinata mitogenome including 13 protein-coding genes, 22 transfer RNA genes, two ribosomal RNA genes for the small and large subunits (rrnS and rrnL) and a putative control region. The ATN initiation codon are used in all protein-coding genes except ATP8 (GTG) and COI (ACTA). The TAN termination codon are used in all protein-coding genes except COI, COII, ND4 and ND5 used the incomplete stop codon T. The assignment of incomplete termination codon on these genes could avoid overlapping nucleotides between their adjacent genes (Gong et al. Citation2012), which is commonly reported in other invertebrates (Masta & Boore Citation2004). And then they would produce functional stop codons via posttranscriptional polyadenylation (Ojala et al. Citation1981). Fourteen overlaps were found between adjacent genes (80 bp in total), the longest is 23 bp located rrnL and trnL1; and including eight intergenic spacers ranged from 1 to 30 bp (58 bp in total), of which only two spacer span longer than 10 bp ().
Figure 1. Phylogenetic tree obtained by the maximum-likelihood (ML) method based on amino acid sequences of ND5 gene.
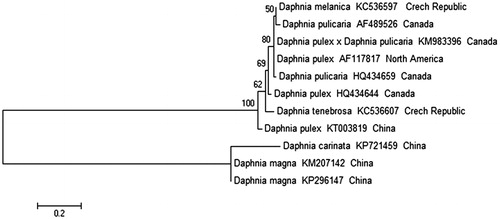
Table 1. Organization of the Chinese Daphnia carinata mitogenome.
All 22 tRNA genes are ranged from 62 bp to 72 bp in size, showing the typical cloverleaf secondary structures except for trnS1, in which its DHU arm simply formed a loop. Generally speaking, the aberrant loop in trnS is a common theme in metazoan mitochondrial genomes. Whether or not the aberrant tRNAs lose their respective functions is still unknown, however, these shortage may be modified by RNA-editing mechanisms (Masta & Boore Citation2004; Li et al. Citation2012). The rrnL and rrnS are 1362 bp and 747 bp, respectively. The rRNA genes with A + T content of 72.69%.
The putative control region is located between rrnS and trnI with a sequenced length of 601 bp, with the A + T content of 72.54%. This region was usually believed to play an important role in the transcription and replication of mitogenome (Clayton Citation1992; Delarbre et al. Citation2001; Delport et al. Citation2002; Yu et al. Citation2013).
The complete mitogenome of the Chinese Daphina carinata reported here is expected to supply more molecular information for further studies of the daphnia phylogeny and for analyses on the taxonomic status of the Cladocera.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Funding information
This work was supported by the National Natural Science Foundation of China (81272377 and 31370470), the Natural Science Foundation of Anhui Province of China (1208085MC45) and the open-ended fund of Anhui Key Laboratory of Plant Resources and Biology (ZYZWSW2014014).
References
- Clayton DA. 1992 . Transcription and replication of animal mitochondrial DNAs. Int Rev Cytol. 141:217–232.
- Delarbre C, Rasmussen A, Arnason U, Gachelin G. 2001 . The complete mitochondrial genome of the hagfish Myxine glutinosa: unique features of the control region. J Mol Evol. 53:634–641.
- Delport W, Ferguson JWH, Bloomer P. 2002. Characterization and evolution of the mitochondrial DNA control region in Hornbills (Bucerotiformes). J Mol Evol. 54:794–806.
- Forró L, Korovchinsky NM, Kotov AA, Petrusek A. 2008. Global diversity of cladocerans (Cladocera; crustacea) in freshwater. Hydrobiologia. 595:177–184.
- Gong YJ, Shi BC, Kang ZJ, Zhang F, Wei SJ. 2012. The complete mitochondrial genome of the oriental fruit moth Grapholita molesta (Busck) (Lepidoptera: Tortricidae). Mol Biol Rep. 39:2893–2900.
- Gu YL, Xu L, Lin QQ, Dumont HJ, Han BP. 2013. The new subspecies of Daphnia: Daphnia similoides sinensis. Ecol Sci. 32:308–312.
- Li H, Liu H, Shi A, Štys P, Zhou X, Cai W. 2012. The Complete Mitochondrial Genome and Novel Gene Arrangement of the Unique-Headed Bug Stenopirates sp. (Hemiptera: Enicocephalidae). Plos One. 7:e29419. doi:10.1371/journal.pone.0029419.
- Masta SE, Boore JL. 2004. The complete mitochnodrial genome sequence of the spider Habronattus oregonensis reveals rearrangement and extremely truncated tRNAs. Mol Biol E. 21:893.
- Miner BE, Knapp RA, Colbourne JK, Pfrender ME. 2013. Evolutionary history of alpine and subalpine Daphnia in western North America. Freshwater Biol. 58:1512–1522.
- Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature. 290:470–474.
- Yu JN, Chung CU, Kwak M. 2013. The complete mitochondrial genome sequence of the Korean hare (Lepus coreanus). Mitochondrial DNA. 26:129–130.
