Abstract
The Arboreal Brown-toothed Shrew (Episoriculus macrurus) belongs to the family Soricidae, and distributes in China, Nepal, India, Myanma, and Vietnam. In this study, the complete mitochondrial genome sequence of E. macrurus was determined. The mitogenome is 16,943 bp in length. Phylogenetic trees were constructed by the methods of Bayesian inference and maximum likelihood. The results showed that in the family of Soricidae, Crocidurinae differentiated earlier than Soricinae. Episoriculus was more close to Nectogale or Neomys rather than Sorex, and Blarinella was close to Sorex. This study contributes to illuminating the taxonomic status of E. macrurus.
The Arboreal Brown-toothed Shrew (Episoriculus macrurus) is a species in the genus Episoriculus of the subfamily Soricinae, family Soricidae. It mainly appears in China (provinces of Yunnan, Sichuan and Tibet), Nepal, India, Myanma and Vietnam (Smith et al. Citation2010; Chakraborty et al. 2013; Jiang et al. Citation2015). This species primarily inhabits temperate broad-leaved forests or scrubs at altitudes of 1500 ∼ 3000 m asl and feeds on invertebrates (Smith et al. Citation2010; Li et al. 2015). Currently, this species was evaluated and listed as Least Concern by IUCN (ver. 3.1), and its population trend is still unknown.
By now, the complete mitochondrial genome about E. macrurus has not been reported, with only partial gene fragments (Cyt b, Rag 2 and CDS) data available (Ohdachi et al. Citation2006; He et al. Citation2010). Here, we sequenced the complete mitochondrial genome and constructed the phylogenetic trees of E. macrurus. The specimen was collected in the Longxi–Hongkou National Nature Reserve, Dujiangyan County, Sichuan, China (latitude: 31.1435° N, longitude: 103.5799° E) and was stored in the Nature Museum of Sichuan University.
The complete mitochondrion of the E. macrurus was 16,943 bp in length (GenBank accession No. KU246040). Similar to other mitochondrial genomes of insectivores, the complete mitochondrial genome of E. macrurus contains 13 typical vertebrate protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes and one control region. The whole base composition of the mitochondrial genome consisted of 33.3% A, 12.8% G, 30.6% T and 23.3% C, with A + T > G + C. The 12S rRNA and 16S rRNA were 960 bp and 1574 bp in length, respectively. The 22 tRNAs ranged from 59 bp (tRNAcys) to 107 bp (tRNAser and tRNAcys) in length, and was distributed through the whole mitochondrial genome. Among the 13 PCGs, ND5 was the longest (1821 bp in length) and ATP8 was the shortest (205 bp in length). The D-loop region was 1484 bp in length and was found between tRNApro and tRNAphe.
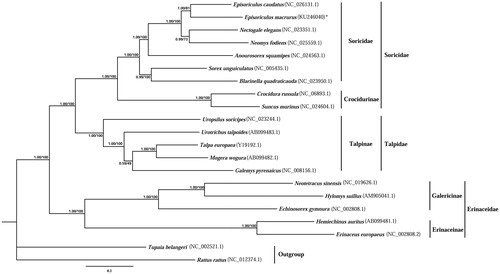
Twenty-one complete mitochondrial genome sequences were used to construct phylogenetic trees based on the concatenated 13 PCGs. Rattus rattus and Tupaia belangeri were chosen as outgroups. Based on the previous studies (Tu et al. Citation2012; Zeng et al. Citation2013; Yan et al. Citation2014), Bayesian inference (BI) and maximum-likelihood (ML) frameworks were used to examine the phylogenetic position of E. macrurus. Both ML and BI analyses generated similar trees (). Our results showed that in the family of Soricidae, Crocidurinae differentiated earlier than Soricinae, which was also supported by He et al. (Citation2012) and Huang et al. (Citation2014). Episoriculus was more close to Nectogale or Neomys rather than Sorex, and Blarinella was close to Sorex. Moreover, E. macrurus and E. caudatus may have close phylogenetic relationship. These two species are sympatric and inhabit similar habitats (Smith et al. Citation2010). This study contributes to illuminating the taxonomic status of E. macruru at the molecular level and essential resource for further research on this species. Further studies are still needed to better understand the phylogenetic relationships within the genus of Episoriculus.
Funding information
This work was supported by the National Natural Science Foundation of China under Grant No. 31200420; Ocean Park Conservation Foundation Hong Kong (OPCFHK) under Grant 2011–2012.
Disclosure statement
The authors declare no conflicts of interest. The authors alone are responsible for the content and writing of this article.
References
- Chakraborty S, Sirinivasalu C, Sirinivasalu B, Pradhan MS, Nameer PO. 2004. Checklist of insect\vores (Mammalia: Insectivora) of South Asia. Zoos Print J. 19:1361–1371.
- He K, Chen JH, Gould GC, Yamaguchi N, Ai HS, Wang YX, Zhang YP, Jiang XL. 2012. An estimation of Erinaceidae phylogeny: a combined analysis approach. PLoS One. 7:e39304.
- He K, Li YJ, Brandley MC, Lin LK, Wang YX, Zhang YP, Jiang XL. 2010. A multi-locus phylogeny of Nectogalini shrews and influences of the paleoclimate on speciation and evolution. Mol Phylogenet Evol. 56:734–746.
- Huang T, Yan CC, Tan Z, Tu FY, Yue BS, Zhang XY. 2014. Complete mitochondrial genome sequence of Nectogale elegans. Mitochondrial DNA. 25:253–254.
- IUCN. 2015. The IUCN Red list of threatened species. Version 2015-4 [Online]. Available from: http://www.iucnredlist.org/.
- Jiang ZG, Ma Y, Wu Y, Wang YX, Zhou KY, Liu SY, Feng ZJ. 2015. China’s Mammal diversity and geographic distribution. Beijing: Science Press.
- Li B, Ran JH, Yue BS, Zhang M, Wu YJ. 2015. Non-volant small mammals in landslides caused by the Wenchuan earthquake in a fragmented forest of Sichuan, China. Pakistan J Zool. 47:535–544.
- Ohdachi SD, Hasegawa M, Iwasa MA, Vogel P, Oshida T, Lin LK, Abe H. 2006. Molecular phylogenetics of soricid shrews (Mammalia) based on mitochondrial cytochrome b gene sequences: with special reference to the Soricinae. J Zool. 270:177–191.
- Smith AT, Xie Y, Hoffmann RS, Lunde D, Mackinnon J, Wilson DE, Wozencraft WC. 2010. A guide to the mammals of China. Princeton, New Jersey: Princeton University Press.
- Tu FY, Fan ZX, Chen SD, Yin YH, Li P, Zhang XY, Yue BS. 2012. The complete mitochondrial genome sequence of the Gracile shrew mole, Uropsilus gracilis (Soricomorpha: Talpidae). Mitochondrial DNA. 23:382–384.
- Yan CC, Zhou Y, Lu L, Tu FY, Huang T, Zhang XY, Yue BS. 2014. Complete mitochondrial genome of Hainan partridge, Arborophila ardens (Galliformes: Phasianidae). Mitochondrial DNA. 25:259–260.
- Zeng T, Tu FY, Ma LL, Yan CC, Yang N, Zhang XY, Yue BS, Ran JH. 2013. Complete mitochondrial genome of blood pheasant (Ithaginis cruentus). Mitochondrial DNA. 24:484–486.