Abstract
We sequenced the complete mitogenome of the Pyrenean frog Rana pyrenaica, which was determined from an Illumina Hi-seq RNAseq run (Illumina Inc., San Diego, CA). The genome is 17,213 bp in size, including 13 protein-coding genes, 21 transfer RNAs, two ribosomal RNAs and a control region. It shows the typical gene order of previously available frog mitogenomes, although it lacks the tRNAPhe. This is the first complete mitogenome described for a Western Palearctic brown frog species.
The Pyrenean frog (Rana pyrenaica) is an Endangered narrowly distributed endemism (Sillero et al. Citation2014). Despite that its sister species Rana temporaria shows a considerable degree of mtDNA genetic variation across its range, including several divergent lineages in the Pyrenees (Vences et al. Citation2013); a preliminary mtDNA study across the range of R. pyrenaica showed a single mutation difference using three mitochondrial genes (Carranza & Arribas Citation2008). This potential lack of genetic variation has important conservation implications for this Endangered species.
RNAseq is becoming a common approach for gathering transcriptome data using Next-Generation Sequencing, being complete mitogenomes a potential output. We explored the use of RNAseq to describe the complete mitogenome of R. pyrenaica, which will benefit future phylogeographic, population genetic and conservation studies.
An adult of R. pyrenaica (Museo Nacional de Ciencias Naturales, Madrid, collection number MNCN 46671) was collected in Uztárroz (42°35′38″N, 0°59′27″W), NE Spain. We extracted RNA from several tissues, which were quantified with Qubit HS and normalized. A RNAseq library was prepared using the NEBNext Ultra RNA kit for Illumina (Illumina Inc., San Diego, CA). Quantification and size estimation were performed on a Bioanalyzer 2100 High Sensitivity DNA chip, and sequenced on 1/2 lane on a Illumina HiSeq (2 × 100 bp pair-end reads). After quality control and trimming with Trimmomatic (v 0.32.2) (Bolger et al. Citation2014), assembly was done with Trinity (v 2.0.6) (Haas et al. Citation2013). Trinity recovered the mitogenome except part of the control region; hence we used this assembly as input for MITObim (Hahn et al. Citation2013) to complete the reconstruction. Genome annotation was done through nucleotide sequence alignments with other ranids. The genome is deposited in GenBank (KU720300).
The complete mitogenome of R. pyrenaica is 17,213 bp in length, including 13 protein-coding genes, two rRNAs, 21 tRNAs and a control region. Gene order, lengths and codon compositions are shown in . The overall base composition of the heavy strand is 27.7% for A, 28.3% for T, 14.9% for G and 29.1% for C, with an A + T bias of 59.9%, similar to other ranid species (Hofman et al. Citation2014, Li et al. Citation2014a, Citation2014b, Ni et al. Citation2015). The genome shows a similar gene organization as other ranids (Kurabayashi et al. Citation2010; Xia et al. Citation2014), but it is the only anuran known so far lacking the tRNAPhe. A maximum-likelihood phylogenetic analysis of ranid frogs based on the available complete mitogenomes () recovers the European R. pyrenaica as the sister taxon to the clade of Asian brown frogs.
Figure 1. Phylogenetic reconstruction of the relationships between ranid frogs, based on available complete mitochondrial genomes except control regions. Maximum-likelihood analyses using a partitioned dataset by codon and gene were performed in RaxML, running for 1000 generations. ML support values are provided above branches. Genbank accession numbers are provided after the species names.
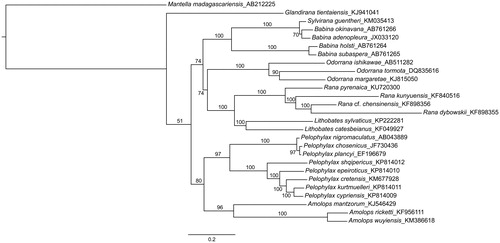
Table 1. Location of features in the mtDNA of R. pyrenaica.
RNAseq has been proven to be a very fast and useful approach to gather complete mitogenomes. Although using common assembly tools like Trinity was not enough to gather the full genome, the combination with MITOBim has performed well to fill the assembly gaps.
Funding information
This work was supported by the Spanish OAPN- Ministry of Environment under Grant 206/2010; Zoo de Barcelona (Ayuntamiento de Barcelona); and the Spanish Ministry of Economy under Grant CGL2013-40924-P. MP was supported by a CNPQ fellowship.
ORCID detail
David R. Vieites http://orcid.org/0000-0001-5551-7419
Acknowledgements
The authors thank the CETA/CIEMAT and the Parque Científico de la UAM for the use of their infrastructures, and the Navarra authorities for collecting permits.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120.
- Carranza S, Arribas O. 2008. Genetic uniformity of Rana pyrenaica Serra-Cobo, 1993 across its distribution range: a preliminary study with mtDNA sequences. Amphibia–Reptilia. 29:579–582.
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protocols. 8:1494–1512.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucl Acids Res. 41:e129.
- Hofman S, Pabijan M, Osikowski A, Szymura JM. 2014. Complete mitochondrial genome of the Greek marsh frog Pelophylax cretensis (Anura, Ranidae). Mitochondrial DNA. 27:1955–1956.
- Kurabayashi A, Yoshikawa N, Sato N, Hayashi Y, Oumi S, Fujii T, Sumida M. 2010. Complete mitochondrial DNA sequence of the endangered frog Odorrana ishikawae (family Ranidae) and unexpected diversity of mt gene arrangements in ranids. Mol Phylogenet Evol. 56:543–553.
- Li J, Lei G, Fu C. 2014a. Complete mitochondrial genomes of two brown frogs, Rana dybowskii and Rana cf. chensinensis (Anura: Ranidae). Mitochondrial DNA. 27:155–156.
- Li J, Yin W, Xia R, Lei G, Fu C. 2014b. Complete mitochondrial genome of a brown frog, Rana kunyuensis (Anura: Ranidae). Mitochondrial DNA. 27:34–35.
- Ni N, Yu D, Storey KB, Zheng R, Zhang J. 2015. The complete mitochondrial genome of Lithobates sylvaticus (Anura: Ranidae). Mitochondrial DNA. doi:10.3109/19401736.2015.1033697.
- Sillero N, Campos J, Bonardi A, Corti C, Creemers R, Crochet PA, Isailović JC, Denoël M, Ficetola GF, Gonçalves J, et al. 2014. Updated distribution and biogeography of amphibians and reptiles of Europe. Amphibia–Reptilia 35:1–31.
- Vences M, Hauswaldt S, Steinfartz S, Rupp O, Goesmann A, Künzel S, Orozco-terWengel P, Vieites DR, Nieto-Román S, Haas S, et al. 2013. Radically different phylogeographies and patterns of genetic variation in two European brown frogs, genus Rana. Mol Phylogen E. 68:657–670.
- Xia Y, Zheng Y, Miura I, Wong PB, Murphy RW, Zeng X. 2014. The evolution of mitochondrial genomes in modern frogs (Neobatrachia): nonadaptive evolution of mitochondrial genome reorganization. BMC Genomics. 15:691.
