Abstract
Taraxacum platycarpum and Taraxacum mongolicum are perennial plants utilized for medicinal purposes in the family Asteraceae. The complete chloroplast genome sequences of the two species were characterized by de novo assembly with whole genome sequencing data. The chloroplast genomes of T. platycarpum and T. mongolicum were 151,307 and 151,451 bp in length, respectively, and showed a typical quadripartite structure. The chloroplast genomes of both species contained the same number of genes, 79 protein-coding genes, 29 tRNA genes and 4 rRNA genes. Phylogenetic analysis indicated that the two Taraxacum species were grouped with T. officinale, all of which showed sister relationship with Lactuca sativa.
Taraxacum platycarpum and Taraxacum mongolicum are perennial plants belonging to the family Asteraceae, the largest families of flowering plants. The genus Taraxacum consists of over 2500 species which is widely grown in the warm temperate areas of the Northern Hemisphere (Richards Citation1973). Taraxacum species have several curative properties including diuretic, choleretic and anti-inflammatory activities and so have long been used for traditional medicinal herbs (Schütz et al. Citation2006). The genus Taraxacum is an evolutionary and taxonomical complex taxa, because of complex hybridity and coexistence of agamosperms, which causes great difficulties in the phylogenetic study of this genus (Kirschner et al. Citation2003; Drábková et al. Citation2009). In this study, we determined the complete chloroplast genome sequences of T. platycarpum and T. mongolicum. These sequences will be valuable genetic resource for further molecular study and phylogenetic analysis of Taraxacum species with other species in the family Asteraceae.
Taraxacum platycarpum and T. mongolicum plants were collected from Chuncheon and Namwon, respectively, in Korea and maintained by Hantaek Botanical Garden (http://www.hantaek.co.kr), Yongin, Korea. Total genomic DNAs were isolated from fresh leaves of each species using a modified cetyltrimethylammonium bromide protocol (Allen et al. Citation2006). Paired-end (PE) libraries were prepared and sequenced using an Illumina MiSeq platform (Illumina, San Diego, CA) by Lab Genomics Inc. (http://www.labgenomics.co.kr), Seongnam, Korea. PE reads of 1.6 and 1.2 Gb were obtained for T. platycarpum and T. mongolicum, respectively, and each species was independently assembled by a CLC de novo genome assembler (v. beta 4.6, CLC Inc., Aarhus, Denmark), as mentioned in Kim et al. (Citation2015a, Citationb). The representative chloroplast contigs were selected, ordered and merged into a single draft sequence by comparing with the previously obtained chloroplast sequence of T. officinale (KU361241) as a reference (Kim et al. Citation2016). The draft sequences were confirmed and manually corrected by PE read mapping. The genes in the chloroplast genomes were annotated using the DOGMA program (Wyman et al. Citation2004) and BLAST searches.
The complete chloroplast genomes of T. platycarpum (GenBank accession KU736960) and T. mongolicum (GenBank accession KU736961) were 151,307 and 151,451 bp in length, respectively. As a typical quadripartite structure found in chloroplast genomes of other angiosperms, the genomes of T. platycarpum and T. mongolicum were separated into four distinct parts, a large single copy (LSC) region (83,922 and 84,052 bp), a short single copy (SSC) region (18,507 and 18,541 bp) and a pair of inverted repeats (IRa and IRb) regions (24,439 and 24,429 bp). A total of 112 genes including 79 protein-coding genes, 29 tRNA genes and 4 rRNA genes were identified in the both chloroplast genomes. The sequence homology of complete chloroplast genomes between T. platycarpum and T. mongolicum was 99.1%.
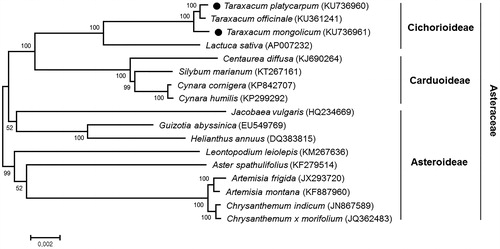
Phylogenetic analysis with other 15 species in the family Asteraceae was carried out based on common 68 chloroplast protein coding sequences, using a maximum likelihood analysis of MEGA 6.0 (Tamura et al. Citation2013) with 1000 bootstrap replicates. The phylogenetic tree showed that T. platycarpum and T. mongolicum were grouped with T. officinale, another Taraxacum species, all of which showed sister relationship with Lactuca sativa (lettuce, a leaf vegetable) in the Cichorioideae subfamily, as expected ().
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of the article. This work was supported by “15172MFDS246” from Ministry of Food and Drug Safety, in 2015, Republic of Korea.
References
- Allen G, Flores-Vergara M, Krasynanski S, Kumar S, Thompson W. 2006. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc. 1:2320–2325.
- Drábková LZ, Kirschner J, Štěpánek J, Záveský L, Vlček Č. 2009. Analysis of nrDNA polymorphism in closely related diploid sexual, tetraploid sexual and polyploid agamospermous species. Plant Syst E. 278:67–85.
- Kim JK, Park JY, Lee YS, Woo SM, Park HS, Lee SC, Kang JH, Lee TJ, Sung SH, Yang TJ. 2016. The complete chloroplast genome sequence of the Taraxacum officinale F.H.Wigg (Asteraceae). Mitochondrial DNA B. [Epub ahead of print].
- Kim K, Lee SC, Lee J, Lee HO, Joh HJ, Kim NH, Park HS, Yang TJ. 2015a. Comprehensive survey of genetic diversity in chloroplast genomes and 45S nrDNAs within Panax ginseng species. PLoS One. 10:e0117159.
- Kim K, Lee SC, Lee J, Yu Y, Yang K, Choi BS, Koh HJ, Waminal NE, Choi HI, Kim NH, et al. 2015b. Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species. Sci Rep. 5:15655.
- Kirschner J, Štěpánek J, Mes TH, Den Nijs JC, Oosterveld P, Štorchová H, Kuperus P. 2003. Principal features of the cpDNA evolution in Taraxacum (Asteraceae, Lactuceae): a conflict with taxonomy. Plant Syst E. 239:231–255.
- Richards AJ. 1973. The origin of Taraxacum agamospecies. Bot J Linn Soc. 66:189–211.
- Schütz K, Carle R, Schieber A. 2006. Taraxacum-a review on its phytochemical and pharmacological profile. J Ethnopharmacol. 107:313–323.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.