Abstract
Currently, the mitochondrial genome of only two species of Sphingidae have been completely sequenced. For the phylogenetic study of Bombycoidea (including Bombycidae, Saturniidae and Sphingidae) using mitochondrial genomes (mitogenomes), more species are required as a basis for future research. In the present study, we sequenced the complete mitogenome of the hawkmoth, Notonagemia analis scribae (Lepidoptera: Sphingidae), to enrich the Sphingidae database. The length of the N. a. scribae genome was 15,303 bp with a typical set of genes (13 protein-coding genes [PCGs], 2 rRNA genes, and 22 tRNA genes), and one major non-coding A + T-rich region. The COI gene had a CGA start codon, which is the start codon for this gene in the majority of lepidopteran species, whereas other PCGs began with ATN codons. A 318-bp A + T-rich region harbored the blocks of conserved sequences that are typically found in lepidopteran insects, excluding a poly-A stretch, which is typically found at the end of the A + T-rich region. Phylogenetic analysis using the 13 PCGs indicated that N. a. scribae grouped together with two within-familial species, Sphinx morio and Manduca sexta, with the highest nodal support both by Bayesian inference and maximum-likelihood methods, forming the Sphingidae monophyletic group.
The lepidopteran family Sphingidae (superfamily Bombycoidea) of hawk moths include ∼1450 species of relatively large-sized moths (van Nieukerken et al. Citation2011). Previously, phylogenetic relationships of three families of Bombycoidea (Bombycidae, Saturniidae and Sphingidae) were studied (Zwick et al. Citation2011), but no mitogenome-based study has been carried out due to limited taxon diversity in the Sphingidae: only Sphinx morio and Manduca sexta are available (Cameron & Whiting Citation2008; Kim et al. Citation2013). Thus, more species from a diverse taxonomic group are necessary for future mitogenome-based phylogenetic studies. Therefore, in the present study, the complete mitogenome of the hawkmoth Notonagemia analis scribae was sequenced, annotated and used for preliminary phylogenetic analysis using the available species of Bombycoidea.
One adult was captured at Mt. Jiri, Jeollabuk-do Province, South Korea (35°20′01′′ N, 127°36′59′′ E). A voucher specimen was deposited at Chonnam National University, Gwangju, Korea. Total DNA was used as the template to amplify three long overlapping fragments (COI-ND4, ND5-lrRNA and lrRNA-COI). Subsequently, 26 short overlapping fragments were amplified using the long fragments as templates. All primers used were Lepidoptera-specific primers designed previously (Kim et al. Citation2012).
The complete mitogenome of N. a. scribae (GenBank accession number KU934302) was 15,303 bp and consisted of two rRNAs, 22 tRNAs, 13 protein-coding genes (PCGs), and 1 major non-coding region (the A + T-rich region). Regarding genome size, N. a. scribae falls between two available sphingids (15,299 bp in S. morio and 15,516 bp in M. sexta) and the gene arrangement is identical to that of other ditrysian Lepidoptera, including Sphingidae, that have the order trnM-trnI-trnQ (where the underline indicates a gene inversion) between the A + T-rich region and ND2 (Cameron & Whiting Citation2008; Kim et al. Citation2011, Citation2013). Twelve of the 13 PCGs started with ATN codons (data not shown), but the COI gene began with CGA (arginine). The 318-bp A + T-rich region displayed several Lepidoptera-specific features, such as the ATAGA motif and adjacent poly-T stretch, and microsatellite-A/T repeat, but a poly-A stretch, which is typically found at the end of the A + T-rich region (upstream of 5′-end of trnM) was found as a mixture of A and T nucleotides (AATATAAATATTA), similar to other sequenced sphingids (data not shown).
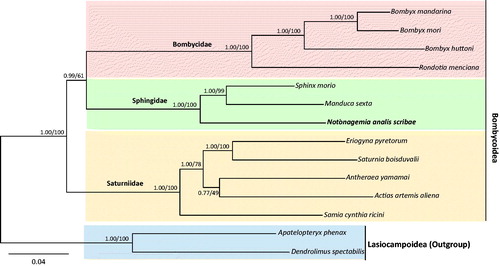
Phylogenetic analysis using nucleotide sequences of 13 PCGs was performed on 12 species in three families of Bombycoidea, including N. a. scribae. Bayesian inference (BI) and maximum-likelihood (ML) methods were performed using the GTR + GAMMA + I model in CIPRES Portal v. 3.1 (Miller et al. Citation2010). The two approaches generated an identical topology, but nodal support varied (). N. a. scribae, grouped together with two within-familial species, S. morio and M. sexta, with the highest nodal support (BI, 1.0; ML, 100%), forming the Sphingidae monophyletic group. Furthermore, each Bombycidae and Saturniidae also formed monophyletic groups with the highest nodal support in both analyses. Although there have been several hypotheses regarding the phylogenetic relationships among bombycoid families (Zwick et al. Citation2011), our limited data supported the sister relationship between Bombycidae and Sphingidae, with the placement of Saturniidae as the basal lineage of the two families. For robust inference of bombycoid families, an increased taxon diversity is required.
Figure 1. Phylogenetic tree for Bombycoidea. Bayesian inference (BI) and maximum-likelihood (ML) methods produced the same topology based on concatenated 13 PCGs. The numbers at each node specify Bayesian posterior probabilities (%) by BI analysis (first value) and bootstrap percentages of 1000 pseudoreplicates by ML analysis (second value). The scale bar indicates the number of substitutions per site. Two species of Lasiocampoidea were utilized as outgroups. GenBank accession numbers are as follows: Notonagemia analis scribae, KU934302 (the present study), Sphinx morio, KC470083 (Kim et al. Citation2013); Manduca sexta, EU286785 (Cameron & Whiting Citation2008); Bombyx mandarina, AB070263 (Yukuhiro et al. Citation2002); Rondotia menciana, KJ647172 (Kim et al. Citation2016); Bombyx huttoni, KP216766 (Peng et al. Citation2015); Bombyx mori, AF149768 (Unpublished); Actias artemis aliena, KF927042 (Park et al. Citation2016); Samia cynthia ricini, JN215366 (Kim et al. Citation2012); Saturnia boisduvalii, EF622227 (Hong et al. Citation2008); Antheraea yamamai, EU726630 (Kim et al. Citation2009); Eriogyna pyretorum, FJ685653 (Jiang et al. Citation2009); Dendrolimus spectabilis, KM244678 (Tang et al. Citation2014); and Apatelopteryx phenax, KJ508055 (Timmermans et al. Citation2014).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
This study was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science, and Technology (2015R1D1A3A03018119).
References
- Cameron SL, Whiting MF. 2008. The complete mitochondrial genome of the tobacco hornworm, Manduca sexta, (Insecta: Lepidoptera: Sphingidae), and an examination of mitochondrial gene variability within butterflies and moths. Gene 408:112–123.
- Hong MY, Lee EM, Jo YH, Park HC, Kim SR, Hwang JS, Jin BR, Kang PD, Kim KG, Han YS, et al. 2008. Complete nucleotide sequence and organization of the mitogenome of the silk moth Caligula boisduvalii (Lepidoptera: Saturniidae) and comparison with other lepidopteran insects. Gene 413:49–57.
- Jiang ST, Hong GY, Yu M, Li N, Yang Y, Liu YQ, Wei ZJ. 2009. Characterization of the complete mitochondrial genome of the giant silkworm moth, Eriogyna pyretorum (Lepidoptera: Saturniidae). Int J Biol Sci. 5:351–365.
- Kim MJ, Choi SW, Kim I. 2013. Complete mitochondrial genome of the larch hawk moth, Sphinx morio (Lepidoptera: Sphingidae). Mitochondrial DNA 24:622–624.
- Kim MJ, Jun J, Kim I. 2016. Complete mitochondrial genome of the mulberry white caterpillar Rondotia menciana (Lepidoptera: Bombycidae). Mitochondrial DNA 27:731–733.
- Kim MJ, Kang AR, Jeong HC, Kim K-G, Kim I. 2011. Reconstructing intraordinal relationships in Lepidoptera using mitochondrial genome data with the description of two newly sequenced lycaenids, Spindasis takanonis and Protantigius superans (Lepidoptera: Lycaenidae). Mol Phylogenet Evol. 61:436–445.
- Kim SR, Kim MI, Hong MY, Kim KY, Kang PD, Hwang JS, Han YS, Jin BR, Kim I. 2009. The complete mitogenome sequence of the Japanese oak silkmoth, Antheraea yamamai (Lepidoptera: Saturniidae). Mol Biol Rep. 36:1871–1880.
- Kim JS, Park JS, Kim MJ, Kang PD, Kim SG, Jin BR, Han YS, Kim I. 2012. Complete nucleotide sequence and organization of the mitochondrial genome of eri-silkworm, Samia cynthia ricini (Lepidoptera: Saturniidae). J Asia Pac Entomol. 15:162–173.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computer Environ Workshop (GCE), New Orleans, LA.
- Park JS, Kim MJ, Kim I. 2016. The complete mitochondrial genome of the moon moth, Actias aliena (Lepidoptera: Saturniidae). Mitochondrial DNA 27:149–150.
- Peng XY, Zhou P, Qiang Y, Qian ZQ. 2015. Characterization of the complete mitochondrial genome of Bombyx huttoni (Lepidoptera: Bombycidae). Mitochondrial DNA Early Online:1–2.
- Tang M, Tan M, Meng G, Yang S, Su X, Liu S, Song W, Li Y, Wu Q, Zhang A, et al. 2014. Multiplex sequencing of pooled mitochondrial genomes-a crucial step toward biodiversity analysis using mito-metagenomics. Nucleic Acids Res. 42:e166.
- Timmermans MJ, Lees DC, Simonsen TJ. 2014. Towards a mitogenomic phylogeny of Lepidoptera. Mol Phylogenet Evol. 79:169–178.
- van Nieukerken EJ, Kaila L, Kitching IJ, Kristensen NP, Lees DC, Minet J, Mitter C, Mutanen M, Regier JC, Simonsen TJ, et al. 2011. Order Lepidoptera Linnaeus, 1758. Zootaxa 3148:212–221.
- Yukuhiro K, Sezutsu H, Itoh M, Shimizu K, Banno Y. 2002. Significant levels of sequence divergence and gene rearrangements have occurred between the mitochondrial genomes of the wild mulberry silkmoth, Bombyx mandarina, and its close relative, the domesticated silkmoth, Bombyx mori. Mol Biol E. 19:1358–1389.
- Zwick A, Regier JC, Mitter C, Cummings MP. 2011. Increased gene sampling yields robust support for higher-level clades within Bombycoidea (Lepidoptera). Syst Entomol. 36:31–43.
