Abstract
Hulless barley (Hordeum vulgar L. var. nudum) is one of the staple foods for Tibetans and an important livestock feed in the Tibetan Plateau. We report the complete mitochondrial genome of Tibetan hulless barely. The complete mitochondrial genome size is 416,675 bp. Hulless barely mitochondrial genome encode 34 protein-coding genes, 19 tRNA genes and three rRNA genes. The mitochondrial genome has 50 forward and palindrome repeats totally. Nucleotide sequence of coding region takes over 13.60%, GC content is 44.33%. The maximum-likelihood (ML) phylogenetic tree based on 11 protein-coding genes common to seven plant mitochondrial genomes using Arabidopsis thaliana as out-group support that hulless barely is close to Triticum species.
Mitochondrial DNA is the extra-nuclear genome in plant cytoplasm except the chloroplast DNA, most likely through the taming of an α-proteobacterial endosymbiont. Lots of evidences support that a single origin of mitochondria for all eukaryotes originated about 1.5 billion years ago (Gupta & Golding Citation1996). Hulless barley as a main food crops and livestock feed more than important of rice where be cultivated in the Tibetan Plateau that high altitude and low temperature. Although the researchers had published the nuclear and chloroplast genome of hulless barely (Zeng et al. Citation2015a, Citation2015b), but mitochondrial genome of hulless barely remains unknown.
Fresh leaf of “Zangqing 320” was used to extract the mitochondrial genome DNA by CTAB protocol (Doyle Citation1987) that has been cultivated in the Tibet Academy of Agricultural and Animal Husbandry Sciences of China. Sequencing was performed on the Hiseq 2000 platform for short insert size libraries (PE500). First, Trimmomatic Version 0.22 (Bolger et al. Citation2014) and Perl scripts were used to data cleaning. Second, compared the clean reads with 75 mitochondrial genomes (ftp://ftp.ncbi.nlm.nih.gov/refseq/release/mitochondrion) by blastn (parameter settings: E-value = le−10, the other parameters are defaults) (Altschul et al. Citation1990). Identity ≥98%, and the ratio of the reads length greater than or equal to 90% was defined as the mitochondrial reads. Thirdly, the mitochondrial reads were assembled by Platnus (Kajitani et al. Citation2014) and GapFiller (Boetzer & Pirovano Citation2012). Five random selected fragments (length=∼1 kB) were verified by PCR method. Finally, the mitochondrial genome was annotated by mitofy (Alverson et al. Citation2010). The GenBank accession number is KU865690.
Repetitive sequence was identified by REPuter (version v3.0, with a minimal length of 50 bp and three mismatches) (Kurtz et al. Citation2001). Reverse and complement type were not detected in hulless barely mitochondrial genome. The totally number of repeats are 50. Forward and palindrome repeats are 29 and 21, respectively. The longest forward and palindrome repeats are 7061 bp and 2366 bp, respectively.
The complete mitochondrial genome of hulless barley has a length of 416,675 bp with a typical circle structure that contains 19 tRNA genes, 34 protein-coding genes and three rRNA genes. Moreover, there are more than one copy of atp8, rRNA and tRNA genes making a total of 88 genes present in hulless barely mitochondrial genome. The percentage of coding region is 13.60%, and GC content is 44.33%.
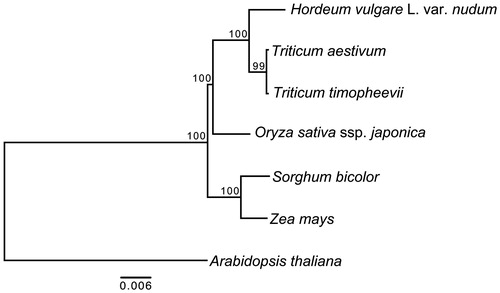
We used 11 protein-coding genes (atp1, atp9, cox1, cox2, cox3, nad4L, nad5, nad6, nad9, rps4 and rps12) common to seven plant mitochondrial genomes for our phylogenetic analysis. Totally 11 protein-coding genes were aligned using the MAFFT version 5 (Katoh et al. Citation2005). Maximum-likelihood analysis was implemented in RAxML version 8.2.4 (Parameters: -# 1000 -m GTRGAMMA –f a) (Stamatakis Citation2006). All other parameters are set as default ().
Figure 1. Maximum-likelihood phylogenetic tree based on 11 protein-coding genes common to seven plant mitochondrial genomes using Arabidopsis thaliana as an out-group. Accession numbers are as follows: Arabidopsis thaliana (NC_001284), Oryza sativa ssp. japonica (NC_011033), Sorghum bicolor (NC_008360), Triticum aestivum (NC_007579), Triticum timopheevii (NC_022714) and Zea mays (NC_007982).
Sequencing the mitochondrial genome of hulless barley will provide essential genomic information for breeding, together with its nuclear and chloroplast genomes is essence to understanding the diversification and evolution of hulless barely.
Funding information
This work was supported by the following funding sources: The National Science and Technology Support Program (2012BAD03B01) and the Tibet Autonomous Region Financial Special Fund (2015CZZX001).
Acknowledgements
The author would thank the editor and reviewers for providing valuable comments on the manuscript.
Disclosure statement
The authors report that they have no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410.
- Alverson AJ, Wei X, Rice DW, Stern DB, Barry K, Palmer JD. 2010. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Mol Biol Evol. 27:1436–1448.
- Boetzer M, Pirovano W. 2012. Toward almost closed genomes with GapFiller. Genome Biol. 13:R56
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120.
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Gupta RS, Golding GB. 1996. The origin of the eukaryotic cell. Trends Biochem Sci. 21:166–171.
- Kajitani R, Toshimoto K, Noguchi H, Toyoda A, Ogura Y, Okuno M, Yabana M, Harada M, Nagayasu E, Maruyama H, et al. 2014. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 24:1384–1395.
- Katoh K, Kuma KI, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucl Acids Res. 33:511–518.
- Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. 2001. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29:4633–4642.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Zeng X, Yuan J, Wang L, Xu J, Tashi N. 2015a. The complete chloroplast genome of Tibetan hulless barley. Mitochondrial DNA 29. [Epub ahead of print]. DOI: 10.3109/19401736.2015.1122765.
- Zeng X, Long H, Wang Z, Zhao S, Tang Y, Huang Z, Tashi N. 2015b. The draft genome of Tibetan hulless barley reveals adaptive patterns to the high stressful Tibetan Plateau. Proc Natl Acad Sci USA. 112:1095–1100.
