Abstract
The complete mitochondrial genome of Gymnocypris potanini firmispinatus was sequenced and compared with others Gymnocypris species. The mitochondrial genome, consisting of 16,680 base pairs (bp), encoded 13 protein-coding genes, 2 ribosomal RNAs, 22 transfer RNAs and a non-coding control region, as those found in other Gymnocypris species. These results can provide useful data for further studies on taxonomic status, molecular systematics and stock evaluation.
Gymnocypris potanini firmispinatus, a subspecies of G. potanini, is only distributed in Jinsha River, the mainstream of Yangtze River, and Yongchun River, a tributary of Lancang River (Yue et al. Citation2000). In recently years, activities like overfishing, dam construction, water pollution and other human interference may exert high pressure on fisheries. Thus, to benefit the effective management of G. potanini firmispinatus resource, some basic biology data including genetic information should be further studied. The mitogenome sequence information may be useful in systematics, resource protection and development.
In the present study, one specimen G. potanini firmispinatus chosen for mitochondrial genome analysis were collected from the upstream of Jinsha River (N′28°04′29.71″, E′102°25′57.90″) (Specimen is stored in Aquaculture Department of Sichuan Agricultural University, number gp20130102). Primers were designed for polymerase chain reaction (PCR) amplification and sequencing on the basis of the mitogenome sequence of G. eckloni Herzensten (GenBank Accession No. JQ0042791) (Qi et al. Citation2013). The complete mt genome of G. potanini firmispinatus was 16,680 bp in length and has been deposited in GenBank with Accession No. KU896808. The mitochondrial genome encoded 13 protein-coding genes, 2 ribosomal RNAs, 22 transfer RNAs and a non-coding control region, as those found in other Gymnocypris species (Qi et al. Citation2013; Qiao et al. Citation2014; Zhang et al. Citation2016). The nucleotide composition of the genome of G. potanini firmispinatus is A 28.4%, T 27.2%, G 18.4%, and C 26.0%, with a low A + T content of 56.6%. Except for the nad6 and eight tRNA genes (tRNA-Gln, tRNA-Aln, tRNA-Asn, tRNA-Lys, tRNA-Tyr, tRNA-SerUCN, tRNA-Glu, and tRNA-Pro) encoded on the light-strand, all others genes were encoded on the heavy-strand. This is a typical gene arrangement conforming to the other Gymnocypris species and vertebrate consensus (Qi et al. Citation2013; Qiao et al. Citation2014).
All genes use ATG as a start codon, except cox1 use GTG, which was also discovered in other Gymnocypris species (Qi et al. Citation2013; Qiao et al. Citation2014). Most open reading frames ended with two types of complete stop codons TAA and TAG, whereas few genes (including cox2, nd4 and cob) had an incomplete stop codon: T––. These results showed that the PCGs are stable among the Gymnocypris species.
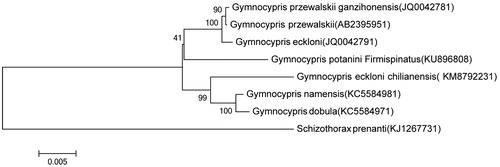
Based on combined nucleotide sequence data of 12 heavy-strand protein-coding genes of G. potanini firmispinatus, and together with the sequences of other Gymnocypris fishes, phylogenetic trees were constructed using the ME methods (). All Gymnocypris species had close relationship, G. przewalskii, G. przewalskii ganzihonensis, and G. eckloni were monophyletic in the trees, which were consistent based on the mitochondrial DNA cytochrome b gene or mitogenoma sequences. In agreement with the findings of previous molecular analyses (Qi et al. Citation2013; Zhang et al. Citation2015), G. dobula and G. namensis also had a close genetic relationship, however, which had a closer distance with G. eckloni chilianensis as the subspecies of G. eckloni (Zhang et al. Citation2015). Thus, the mitochondrial genome data and phylogenetic analysis of the G. potanini firmispinatus can enrich the evolution research of Gymnocypris.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
References
- Qi D, Chao Y, Zhao L, Shen Z, Wang G. 2013. Complete mitochondrial genomes of two relatively closed species from Gymnocypris (Cypriniformes: Cyprinidae): genome characterization and phylogenetic considerations. Mitochondrial DNA. 24:260–262.
- Qiao H, Cheng Q, Chen Y. 2014. Characterization of the complete mitochondrial genome of Gymnocypris namensis (Cypriniformes: Cyprinidae). Mitochondrial DNA. 25:17–18.
- Yue P, Shan X, Lin R. 2000. Fauna Sinica: Osteichthyes Cypriniformes III. Beijing (China): Science Press.
- Zhang J, Chen Z, Zhou C, Kong X. 2016. Molecular phylogeny of the subfamily Schizothoracinae (Teleostei: Cypriniformes: Cyprinidae) inferred from complete mitochondrial genomes. Biochem Syst and Ecol. 64:6–13.
- Zhang JP, Liu Z, Zhang B, Yin XY, Wang L, Shi HN, Kang YJ. 2015. Genetic diversity and taxonomic status of Gymnocypris chilianensis based on the mitochondrial DNA cytochrome b gene. Genet Mol Res. 14:9253–9260.