Abstract
Rufous-breasted Accentor (Prunella strophiata) is a small-sized bunting with an extremely geographical range in the world. Here, the complete mitochondrial genome of P. strophiata (16,830 bp in length) has been analyzed for building the database. Similar to the typical mtDNA of vertebrates, it contained 37 genes (13 protein-coding genes, two rRNA genes and 22 tRNA genes) and a non-coding region (D-loop). Overall base composition of the complete mitochondrial DNA is A (30.3%), G (14.7%), C (31.0%) and T (24.0%), the percentage of A and T (54.3%) is higher than G and C (45.7%). All the genes in P. strophiata were distributed on the H-strand, except for the ND6 subunit gene and 9 tRNA genes which were encoded on the L-strand. The phylogenetic relationships of 12 Passeriformes species were reconstructed based on the complete mtDNA sequences using the Bayesian inference method.
Rufous-breasted Accentor (Prunella strophiata) is a small-sized passerine bird, which is reported to be locally frequent in Pakistan and China, common in northern India, fairly common in Nepal and common in Bhutan (Grimmett et al. Citation1998; Zheng Citation2002). The population is suspected to be stable in the absence of evidence for any declines or substantial threats. Currently, it has been categorized as Least Concern (LC) by IUCN. Up to now, only partial sequence of the mitochondrial DNA of Rufous-breasted Accentor is available. Considering the small size, maternal inheritance, accelerated rate of mutation compared to the nuclear DNA and little or no recombination which was useful for phylogenetic relationships at several taxonomic levels (Brown et al. Citation1979; Ballard & Whitlock Citation2004), we analyze the complete mitochondrial genome of P. strophiata for preserving the commercial and biodiversity value and reconstructing the phylogeny.
An adult Rufous-breasted Accentor was collected from Shaanxi Foping National Nature Reserve, which is numbered 20151201 and stored at the museum in the School of Life Sciences, Anhui University, China. The genomic DNA was extracted from the muscle tissue using standard phenol-chloroform methods (Sambrook & Russell Citation2001). The complete mtDNA sequences of P. strophiata (accession no. KU975800) is 16,830 bp in length which was sequenced by High-throughput sequencing technology. It encode 37 genes totally containing 13 protein-coding genes (PCGs) and 22 tRNA genes, two rRNA genes and non-coding region (D-loop). All the genes in P. strophiata were distributed on the H-strand, except for the ND6 subunit gene and nine tRNA genes which were encoded on the L-strand. Overall base composition of the complete mitochondrial DNA is A (30.3%), G (14.7%), C (31.0%) and T (24.0%), which indicated a strong AT bias (Shadel & Clayton Citation1997).
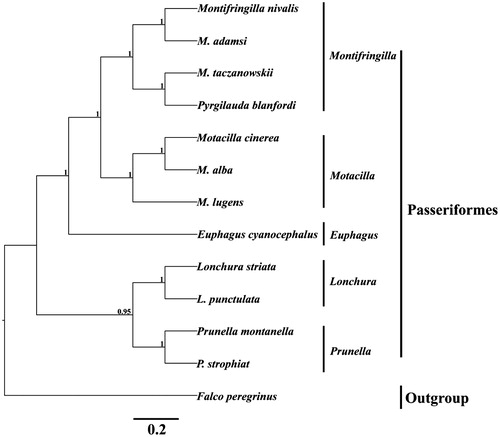
In order to convince the mitochondrial sequence obtained in this study, we selected 12 species from the Passeriformes order to reconstruct the phylogenetic tree with Bayesian inference (BI) method using the MrBayes version 3.1 (Huelsenbeck & Ronquist Citation2001). The best-fitting nucleotide substitution model (GTR + I + G) was selected by MrModeltest version 2.1 (Nylander Citation2004). The Markov Chain Monte Carlo (MCMC) was run with four chains for 1,000,000 generations until the average standard deviation (SD) of split frequencies reached a value less than 0.01. Bayesian posterior probabilities calculated from the sample points after the MCMC algorithm had started to converge (Zhan & Fu Citation2011). The phylogenetic tree is classified into five clades (). The first lineage, containing four species (Montifringilla nivalis, M. adamsi, M. taczanowski and Pyrgilauda blanfordi), belongs to genus Montifringilla. The second lineage, genus Motacilla, includes three species (Motacilla cinerea, M. alba and M. lugens). The third group, only contains Euphagus cyanocephalus. The forth group, genus Lonchura, contains Lonchura striata and L. punctulata. The last group, genus Prunella, sister group to Lonchura. We expect that the present result can contribute to construct molecular identification of this species and be helpful to explore the phylogeny of Passeriformes.
Figure 1. Inferred phylogenetic relationships of 12 Anatidae species were reconstructed based on the complete mtDNA sequences using Bayesian inference (BI). Numbers at each node indicate percentages of Bayesian posterior probabilities (BPPs). GenBank accession numbers for the published sequences are NC_025911 (Montifringilla nivalis), NC_025913 (Montifringilla adamsi), NC_025914 (Montifringilla taczanowskii), NC_025912 (Pyrgilauda blanfordi), NC_027933 (Motacilla cinerea), NC_029229 (Motacilla alba), KU246035 (Motacilla lugens), NC_018827 (Euphagus cyanocephalus), NC_029475 (Lonchura striata), NC_028036 (Lonchura punctulata), NC_027284 (Prunella montanella), KU975800 (Prunella strophiat) and NC_000878 (Falco peregrinus).
Acknowledgements
The authors are especially grateful to Wei Wei for their laboratory assistance and data analysis.
Disclosure statement
Research supported by the Foundation for Graduate Student Academic Innovation Research Project of Anhui University (yqh100112). The authors report that they have no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Ballard JWO, Whitlock MC. 2004. The incomplete natural history of mitochondria. Mol Ecol. 13:729–744.
- Brown WM, George M, Wilson AC. 1979. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci USA. 76:1967–1971.
- Grimmett R, Inskipp C, Inskipp T. 1998. Birds of the Indian subcontinent. London: Christopher Helm.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17:754–755.
- Nylander J. 2004. MrModeltest v2. Program distributed by the author. Uppsala: Uppsala University.
- Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. New York: Cold Spring Harbour Laboratory Press.
- Shadel GS, Clayton DA. 1997. Mitochondrial DNA maintenance in vertebrates. Annu Rev Biochem. 66:409–435.
- Zhan A, Fu J. 2011. Past and present: phylogeography of the Bufo gargarizans species complex inferred from multi-loci allele sequence and frequency data. Mol Phylogenet Evol. 61:136–148.
- Zheng GM. 2002. A checklist on the classification and distribution of the birds of china. Beijing: Science Press.
